An Introduction to Teaching with Biomolecular Visualization
Matthew Flagg and Henry V. Jakubowski
Navigating this Book: To progress between chapters, use the left and right arrow keys on your keyboard, or the “Previous” and “Next” buttons on the bar at the bottom of each page. To view a list of chapters and navigate, use the menu at the top of the page. The top menu bar navigation is a useful tool for jumping between book sections while working through the activities for a specific modeling program.
The Goal of This Open Educational Resource
Molecular visualization (MV) allows instructors and students to explore and analyze a universe of biological structures, ranging from small molecules to complex macromolecular assemblies. Unfortunately, effective use of MV requires surmounting the learning curves of powerful but not-always-intuitive modeling programs, such as PyMOL, ChimeraX, iCn3D, and Mol*. How can you efficiently teach yourself and your students to use those tools? In this textbook, we aim to train instructors in the fundamentals of MV, and we highlight the pros and cons of different MV tools. This text will also provide materials that faculty could adapt to their classroom needs.
Why Bring Molecular Visualization into Your Classroom?
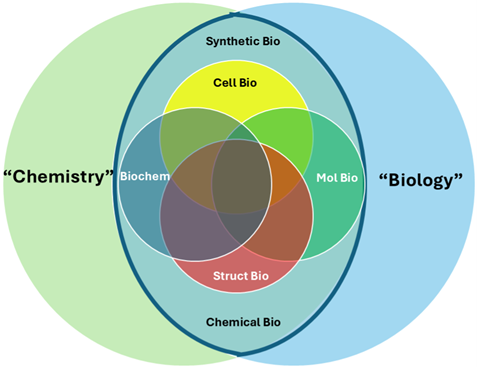
Chemistry and biology present different challenges. The overlap of these fields into disciplines such as biochemistry, molecular and cell biology, structural biology, and their newer variants, synthetic and chemical biology, presents additional challenges to faculty and student understandings (Figure 1). What unites these fields is the threshold concept of biomolecular structure and the emergence of function from it (Loertscher et al., 2014). The use of MV tools provide a means to visualize and thus aid in understanding biomolecular structure and function, unifying teaching across courses while lowering disciplinary barriers. It can connect our disciplines and provide students with a standard and coherent framework as they try to understand the relationships between structure and function in biology and chemistry.
An example illustrates the power of MV (Figure 2, left): An antibody (yellow) binds to a spike protein (magenta) on the surface of the SARS-CoV-2 virus. Binding to the spike protein effectively blocks the virus from entering cells and prevents infection. This is the biomolecular function, dictated by biomolecular structure, in all its elegant simplicity — and complexity. Look closer at the binding interface (Figure 2, right), and you enter the exquisite world of biomolecular interactions, enabled by eons of evolution and modified by adaptation. MV provides a gateway to a deeper understanding of structural interactions in the molecular world, potentially unlocking new medicines and biological catalysts that can improve our lives.
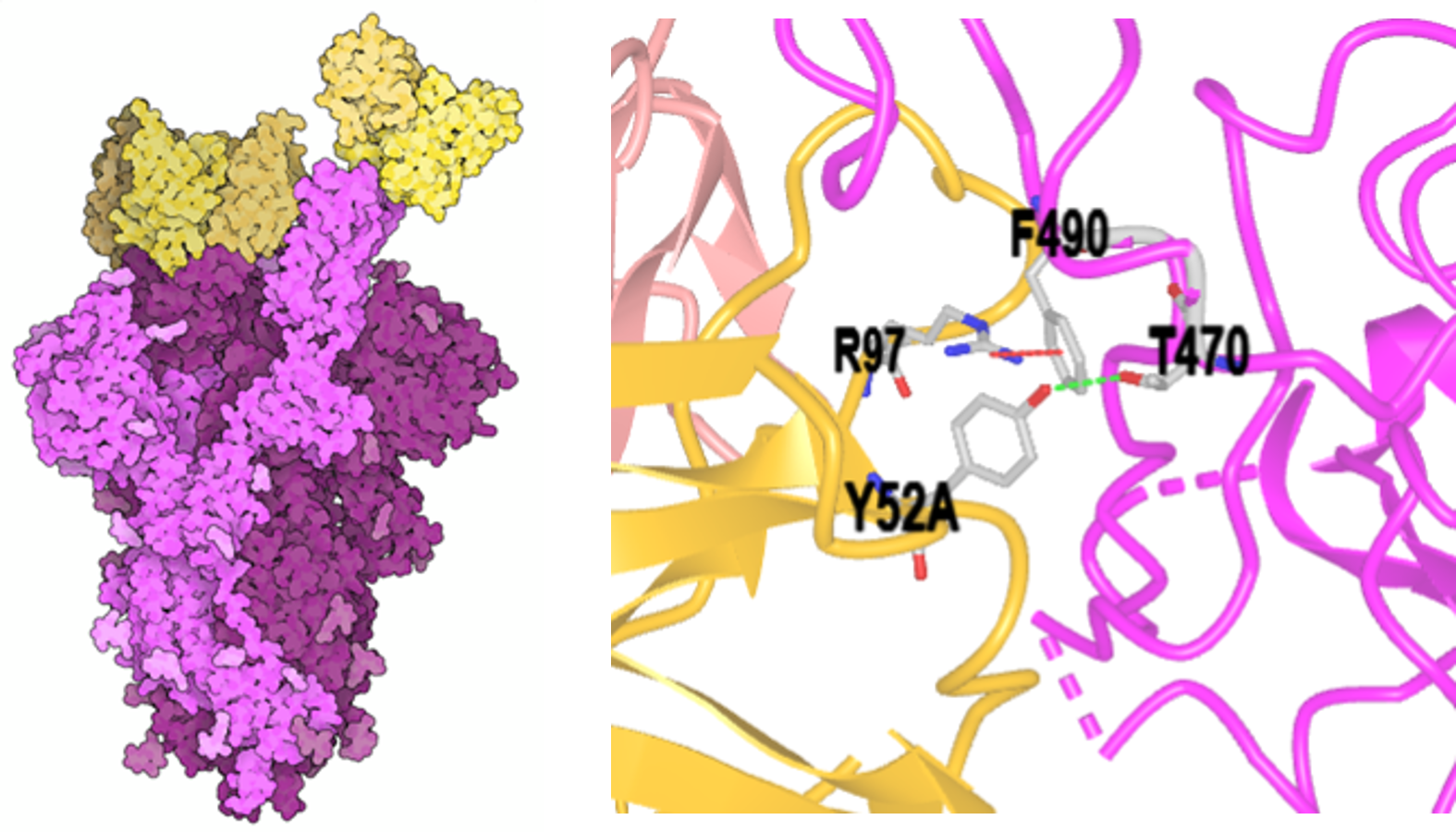
However, people interpret visual images differently based on their training and experiences. To understand the figures above, students must recognize that different renderings of a given structure are explicitly used to highlight different aspects of structure and derived function. Explicit training in biomolecular visualization is needed to bridge how biologists, chemists, students, and instructors understand macromolecular structures.
When students master MV, they can make broad connections between the seemingly disparate chemistry, biochemistry, and biology disciplines. For instance, they can begin with the structure of a ligand and recognize the spatial distribution of charges across the molecule. Then, they can inspect how a binding pocket presents corresponding charges and a corresponding shape that fits the ligand at the atomic level. Students can use the tools described herein to produce images of these interactions, demonstrating a deep and grounded understanding. Chemical logic transitions into biochemical logic seamlessly with the possibility for active engagement.
References
Goodsell, D. (2021) Molecule of the Month: SARS-CoV-2 Spike and Antibodies. http://doi.org/10.2210/rcsb_pdb/mom_2021_4
Loertscher, J., Green, D., Lewis, J. E., Lin, S., & Minderhout, V. (2014). Identification of threshold concepts for biochemistry. CBE life sciences education, 13(3), 516–528. https://doi.org/10.1187/cbe.14-04-0066
Jakubowski, H. (2025) Interactions of Spike Protein and Antibody PDB ID: 7K8T https://www.ncbi.nlm.nih.gov/Structure/icn3d/share.html?9nBUrwnUAS86g7o89
