XII.B. iCn3D Mutagenesis
Henry V. Jakubowski and James Nolan
Overview: This activity demonstrates how to manually modify an amino acid residue to simulate protein mutagenesis.
Outcome: The user will be able to change the identity of an amino acid residue in order to visualize the impact of mutagenesis on the protein of interest.
Time to complete: 30 minutes (15 minutes per example).
Modeling Skills
- Changing the identity of an amino acid residue using the mutagenesis tool
- Visualizing the impact of mutagenesis on the interactions in a protein
About the Models
PDB ID: 4LTV
Protein: Epi-isozizaene synthase (EIZS)
Activity: Cyclizes the isoprenoid precursor, farnesyl diphosphate (FPP) to form the tricyclic terpene epi-isozizaene. Epi-isozizaene is an intermediate in the biosynthesis of the natural product albaflavenoid.
Description: Single chain, no ligands bound in the active site, two sulfate ions
Note: We will be using the commands to change the identity of a specific amino acid residue in the enzyme. The amino acid residue, F96, that is mutated in this exercise is on the inner surface of the active site pocket. Mutagenesis experiments (Aaron et al, 2010, Li. et al., 2014) have shown that when this residue is mutated, the change in the contour and electrostatics of the active site pocket result in a dramatic change in the identity of isoprenoid products formed. The F96A mutation changes main product of the enzyme from epi-isozizaene to β–farnesene, and the F96W mutation changes the main product from epi-isozizaene to (+)-zizaene.
Steps
Substitution (mutagenesis) of an amino acid residue with rotamer optimization using scap
-
- Load 4LTV
a) Open iCn3D
b) In: List of PDB, MMDB, or AlphaFold UniProt structures, type 4LTV
c) Click enter or “Load Biological Unit”.
- In the dropdown menu: Analysis → Mutation.
a) Input the desired mutation F96W this way: 4LTV_A_96_W, where A is the A chain, 96 is the aa #, and W is the new mutated amino acid residue Trp. To view that mutant and wild type, there are 3 different possible analysis methods: 3D with scap, Interactions, and PDB. We will begin with scap. The scap program first iteratively samples all sidechain rotamers until convergence. The final lowest-energy conformation among all the conformations starting from the initial three conformations is then minimized by refining the side-chain conformation with rotation on each bond in the sidechain to search for lower energy conformations around the rotamer.
b) Check Show Mutation in New Page. - Click 3d with scrap. Choose OK to go back and forth between structures using the “a” key. Input “a” several times to toggle between the WT structure, 4LTV and the mutant, 4LTV2. Last “a” should end on the WT, 4LTV.
- In the dropdown menu: Select → Defined Sets.
- Click 4LTV_A, to select the WT protein.
- In the dropdown menu: Color → Unicolor → Cyan → Cyan.
- Input “a” to toggle to the mutant, 4LTV2.
- In the Select Sets window, click 4LTV2_A, to select the mutant.
- In the dropdown menu: Color → Unicolor → Orange → Orange.
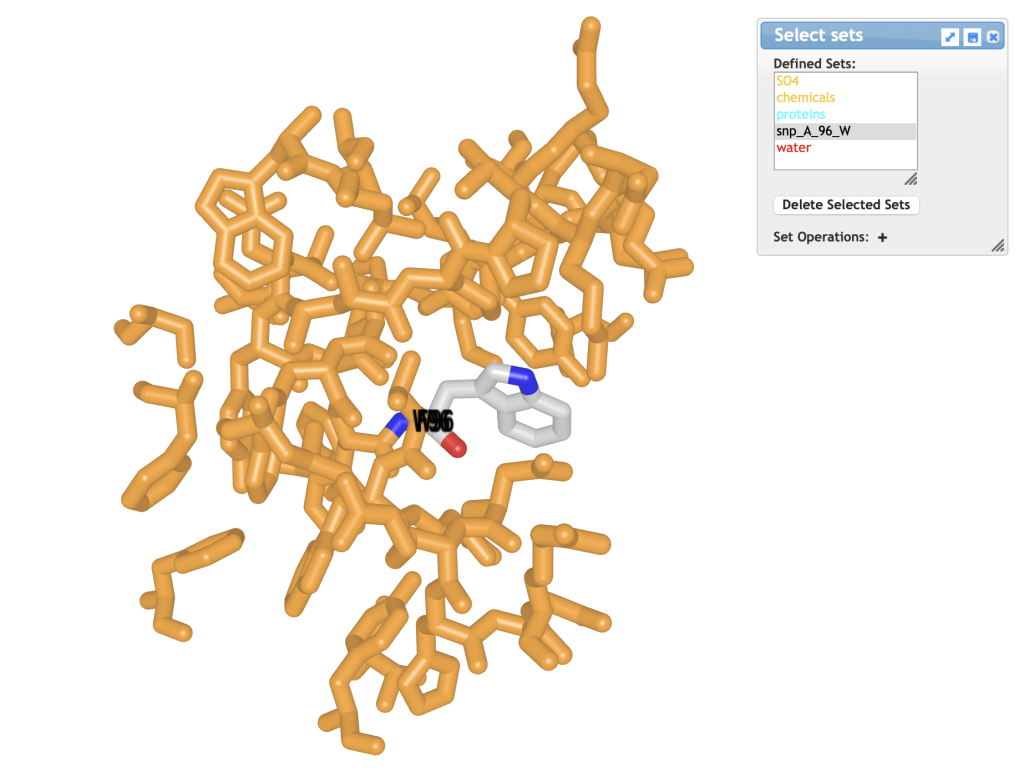
Figure 1. Structure of F96W mutant of 4LTV protein. The mutant Trp residue is colored in CPK colors. - In the Select Sets window, choose [snp_A_96_W]; this selects only the mutated Trp residue (see window at top of Figure 1).
- In the dropdown menu: Color → Atom
- With the Trp highlighted yellow: In the dropdown menu: Analysis → Label → Per Reside and Number. This labels just the mutant W.
- In the dropdown menu: Analysis → Label Scale → 2.
- In the dropdown menu: Style → Background → Transparent. View should be similar to Figure 1; toggle between mutant and wild type by inputting“a”.
- In the dropdown menu: File → Share Link → Lifelong Short URL.
- Here is a URL for an example: https://www.ncbi.nlm.nih.gov/Structure/icn3d/share2.html?ff4ea070522d45d7c35637d10d8440ff .
Substitution (mutagenesis) of an amino acid residue with analysis of interactions
-
-
- Load 4LTV
a) Open iCn3D
b) In List of PDB, MMDB, or AlphaFold UniProt structures, type 4LTV
c) Click enter or “load biological unit”.
- In the dropdown menu: Analysis → Mutation.
a) This time we will input an F96A (alanine) mutation this way: 4LTV_A_96_A, where the first A is the A chain, 96 is the aa #, and the second A is the new mutated amino acid residue Ala. This time we will look at effects on interactions between residues.
b) Check Show Mutation in New Page. - Click Interactions. Choose OK to go back and forth between structures using the “a” key. Input “a” several times to toggle between the WT structure, 4LTV and the mutant, 4LTV2. Last “a” should end on the WT, 4LTV.
- In the dropdown menu: Style → Background → Transparent.
- Optional: Change colors of each chain and atoms, if desired, by repeating steps 6 – 9 above in the first exercise.
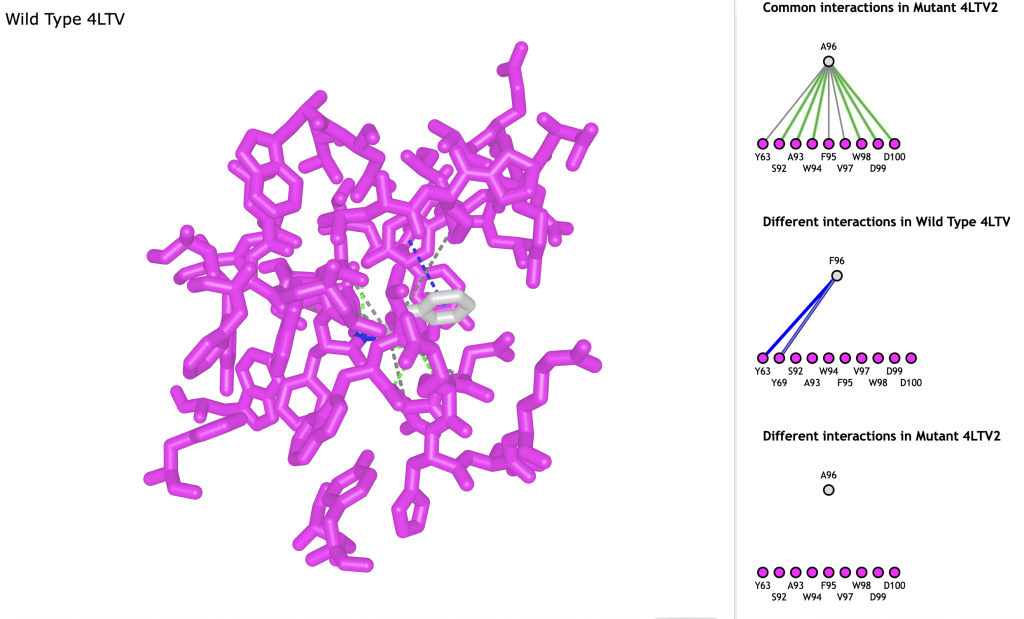
Figure 2. Interaction analysis of wild type 4LTV protein. All amino acids are rendered in magenta, except Phe-96. Window at right shows interactions between amino acids: pi-stacking interaction are shown as blue dashed lines, H-bonds are shown as green dashed lines, additional contacts are shown as gray dashed lines. - A new window appears after step 3: The lines show interactions between pairs of residues. Read through the list and scroll down to the next-to-last description: Different interactions in Wild Type 4LTV2.
- Click the dark blue line connecting the two residues to highlight the interaction found in the wild type but missing in the mutant. Input “a” to toggle between wild type and mutant to see the interactions lost in the mutant.
- In the dropdown menu: File → Share Link → Lifelong Short URL.
- Here is a URL for an example: https://www.ncbi.nlm.nih.gov/Structure/icn3d/share2.html?c0c981caf9f1a9a45b7ed2dcfb095c83 .
- Clicking any line in the interactions window selects the interaction amino acids and highlights them in yellow. Clicking the line again zooms in on the interacting amino acids. Individual amino acids can be selected by clicking on a dot instead of the line (Figure 3).
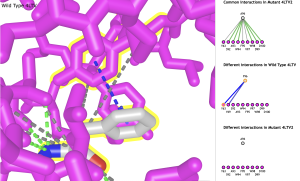
Figure 3. Zoomed in view of interaction between Phe-96 and Tyr-63.
-
