XI.D. PyMOL Superimposition
Roderico Acevedo and Kristen Procko
Overview: This activity demonstrates how to align two objects in the modeling program.
Outcome: The user will be able to superimpose two similar macromolecules to examine commonalities and differences.
Time to complete: 5 minutes
Modeling Skills
- Superimposing objects
About the Models
PDB ID: 1xww & 5pnt (different isozymes)
Protein: Low molecular weight protein tyrosine phosphatase
Activity: Hydrolyzes Tyr-OPO32- phosphoester bond
Description: Single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Note: We will be using the command super for superimposing two isozymes. PyMOL has three different commands to accomplish this task: align, super, and cealign. Briefly, align is recommended for structures that have high sequence similarity, while super is a sequence-independent command useful for structurally similar macromolecules. The command cealign is recommended for low sequence identity macromolecules. For more detailed information, please consult the PyMOL wiki page on superimposition.
Steps
- Fetch the two structures. In the command line, type: fetch 1xww 5pnt, type=pdb1
- Apply the superimpose command. In the command line, type: super 1xww, 5pnt
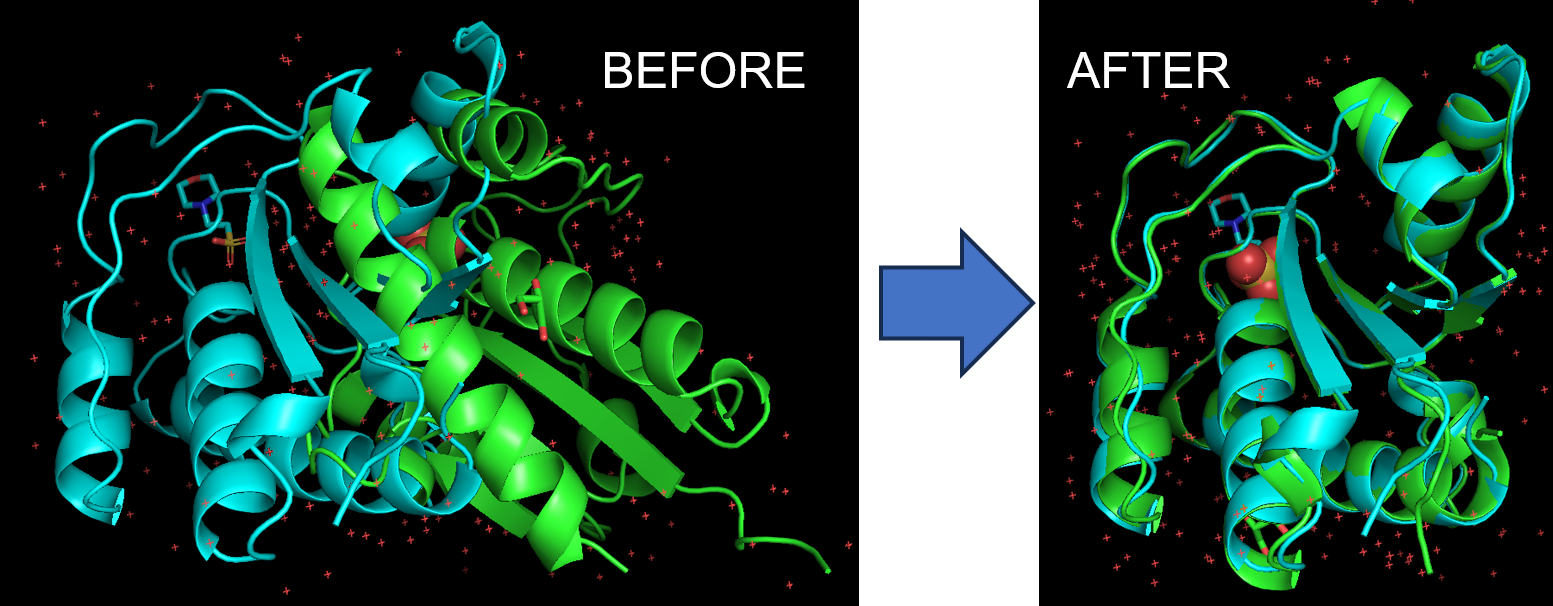
Note: The first structure moves to superimpose onto the second structure. If you wish for 1xww to remain in the same position, type: super 5pnt, 1xww
- Select a 5 Å zone around MES (2-(N-Morpholino)-ethanesulfonic acid) and display those residues as sticks. It may be helpful to hide the cartoon backbone of the two proteins.
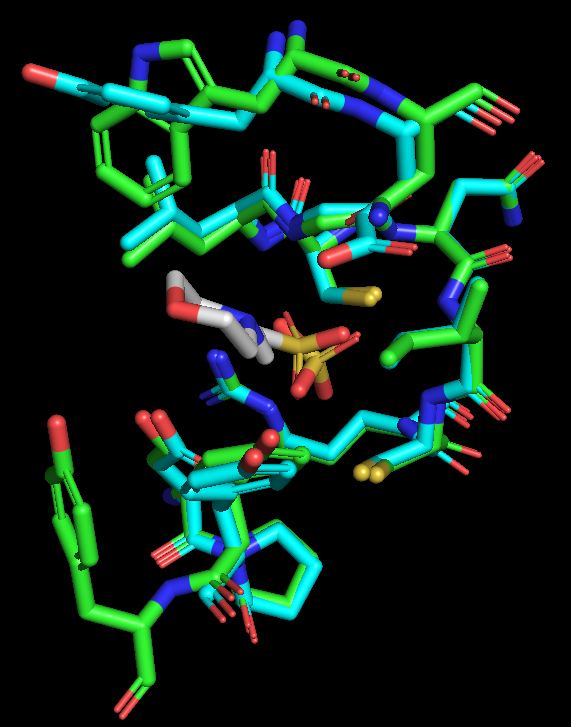
Figure 2: Only active site residues are shown, as colored sticks.
What differences can you see between the active sites of the proteins?
Video Tutorial
Here is a video on superimposition in PyMOL (especially for when the super command doesn’t work well).
Click here to go to Chapter XII: Mutagenesis
