III.D. PyMOL Molecular Surface/Properties
Roderico Acevedo and Kristen Procko
Overview: This chapter describes the application of coloring to help display molecular properties.
Outcome: The user will be able to display the molecular surface of a structure, apply a chemical property-based coloring scheme, and change transparency.
Time to complete: 10 minutes
Modeling Skills
- Display the Molecular Surface of Protein
- Molecular with different colors
- Electrostatic potential
- Hydrophobicity
About the Model
PDB ID: 1XWW
Protein: Low molecular weight protein tyrosine phosphatase
Activity: Hydrolyzes Tyr-OPO32- phosphoester bond
Description: Single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
Load the Structure
- In the command line, type: fetch 1XWW, type=pdb1
Generate Molecular Surfaces and Surface Transparency
- In the names/object panel, beside 1xww, click: S > Surface
- You may change the color of the surface, in the names/object panel, beside 1xww, click C and choose a color from the dropdown menu
Note: Changing colors using the names/object panel will change the color of all the atoms. For example, choosing green in the dropdown will change all atoms green and when surface is hidden, the underlying cartoon will also be green.
- To change the surface color independently, we use the command line. In the command line type: set surface_color, (color), 1xww
where (color) is the color of your choosing. You can use the dropdown menu in C to view a list of color names.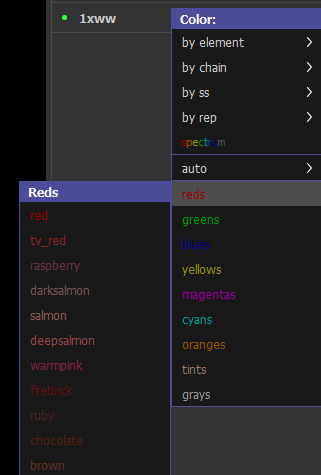
Figure 1: Color choices available in the color menu. - To make the surface semitransparent, in the command line type: set transparency, 0.5
where 0.5 is 50% transparent.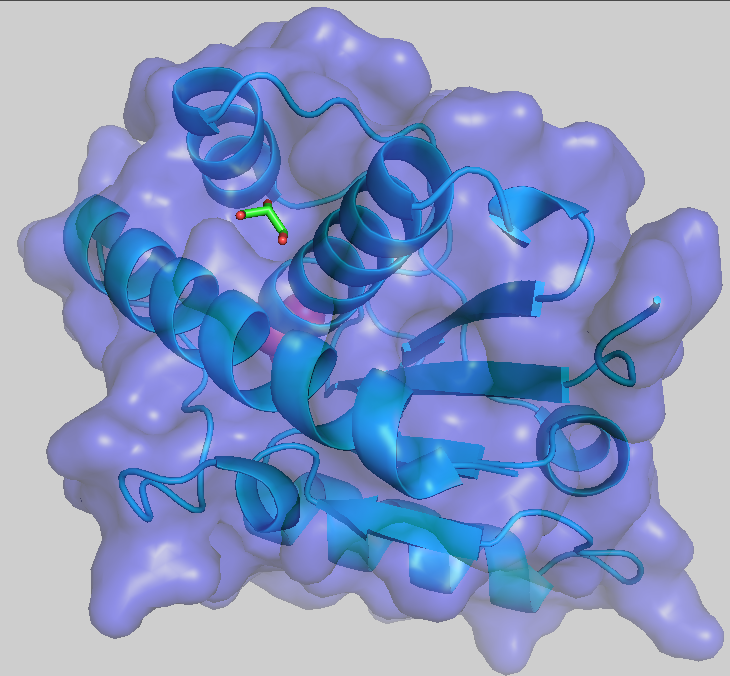
Figure 2: Results of making surface semi-transparent.
Generate an Electrostatic Potential Surface
- In the dropdown menus, click Plugin → APBS electrostatics.
- In the pop-up box, click “Run”. A black pop-up will open and begin running calculations (this may require you to run PyMOL as administrator). A successful run will end with a small pop-up that says: “Finished with Success”.
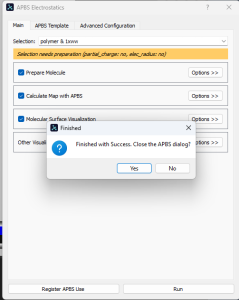
- APBS will calculate the coulombic potential and color the surface according to the calculated charge. Red = negative, white = neutral, blue = positive.
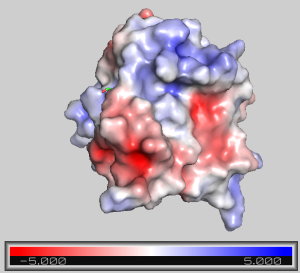
- As above, this surface may be made semitransparent. To undo this, in the command line type: set transparency, 0
Note: The generated surface is overlayed onto any previously made surfaces. Make sure to hide any other surface renderings that may be turned on.
Generate a Hydrophobicity Surface
Note: PyMOL does not have an in-house plugin that will calculate hydrophobicity and plot its values onto a surface. There are scripts, such as color_h.py that will color protein molecules according to the Eisenberg hydrophobicity scale.
Click here to go to Chapter IV: Loading Structures
