VII.D. PyMOL 5Å Selection
Roderico Acevedo and Kristen Procko
Overview: This activity demonstrates how to use one part of a macromolecular structure to select nearby objects.
Outcome: The user will be able to select a specific part of a macromolecule of interest, display surrounding residues that are within range to interact with the selection, and save these selections.
Time to complete: 15 minutes
Modeling Skills
- Selecting objects using the command line
- Selecting groups within 5 Angstroms
About the Model
PDB ID: 1xww
Protein: Low molecular weight protein tyrosine phosphatase
Activity: Hydrolyzes Tyr-OPO32- phosphoester bond
Description: Single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
Load the Structure
- Reload 1xww using the command line. Type: fetch 1xww, type=pdb1
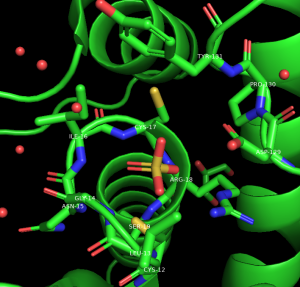
Selecting Objects within a Distance
- Hide the waters in the structure
- Select the sulfate using the command line. Type:
select sulfate, resn SO4 - Click on sulfate → S → sticks
- Use this selection to define the area around the ligands by first duplicating it, click on sulfate → A → Duplicate. Then, click on sel01 → A → rename selection
- Using the keyboard, delete the letters “se101” and type: active
- Modify this selection to show residues within 5Å. In the names/object panel. Click: active → A → Modify → Around → residues within 5Å. Then, to show these residues as sticks, click: active → S → Sticks.
- To label the residues, click L → Residues
- To keep the labels on top, in the command line type: set float_labels, on
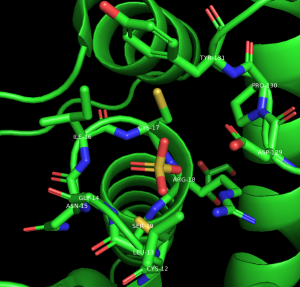
Show Active Site Water
-
In the names/object panel, beside active: A → Duplicate.
-
To rename the selection, click on Sel02 → A → Rename Selection. Delete the letters in the renaming menu that appears in the top right of the structure viewer, and type: active_water
-
To adjust the new selection to contain active site water molecules, click: active_water → A → Modify → Around → Atoms Within 4 Angstroms.
-
To modify this further and limit to water molecules, click on Active_water → A → Modify → Restrict → To Solvent.
Note: The GUI allows selection within 4Å; line commands allow selection of a more appropriate distance of 3.3Å for hydrogen bonding water molecules. The van der Waals radii of the spheres cannot be set in the GUI, but the “ball and stick” selection is close to 0.5Å.
Use Commands to Show Active Site Water
- To reinitialize PyMOL in the command line, type: reinit
-
Reload 1xww using the command line. Type: fetch 1xww, type=pdb1
-
Hide the waters in the structure. In the command line, type: hide nonbonded
- Select the sulfate using the command line. Type:
select sulfate, resn SO4
show sticks, sulfate
hide spheres, sulfate
- Use the command line to select the active site this time:
sele active, byres all within 5 of sulfate
show sticks, active
- Select any water molecules near the active site, using the command line, by typing each line of the following code:
select active_water, ((active)around 3.3) and (resn HOH)
show spheres, active_water
alter active_water, vdw=0.5
rebuild
- Label the active site residues with their names to identify interacting residue. In the names/object panel, click: active → L → residues
- In the command line, type: set float_labels, on
Click here to go to Chapter VIII: Molecular Interactions