VII.A. ChimeraX 5Å Selection
Walter Novak and Josh Beckham
Overview: This activity demonstrates how use one part of a macromolecular structure to select nearby objects.
Outcome: The user will be able to select a specific part of a macromolecule of interest, display surrounding residues that are within range to interact with the selection, and save these selections.
Time to complete: 15 minutes
Modeling Skills
- Selecting objects using the command line
- Selecting groups within 5 Angstroms
About the Model
PDB ID: 1xww
Protein: Low molecular weight protein tyrosine phosphatase
Activity: hydrolyzes Tyr-OPO32- phosphoester bond
Description: single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
Load Structure
-
Load 1xww using the command line. Type: open 1xww
Selecting Objects within a Distance
-
Hide all the residues, keeping only the cartoon. Ensure “Home” is selected in the toolbar selector, and click “Hide” in the Atoms toolbar.
-
Select the sulfate using the command line. Type: sel :SO4
-
To select a region within 5Å of the sulfate, use the dropdown menu: Select → Zone…
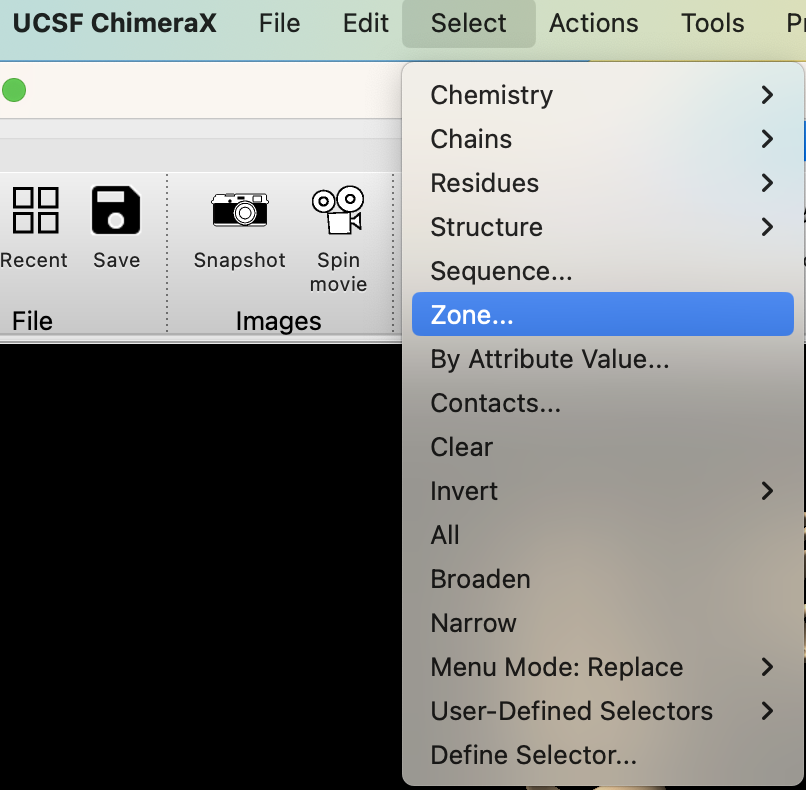
-
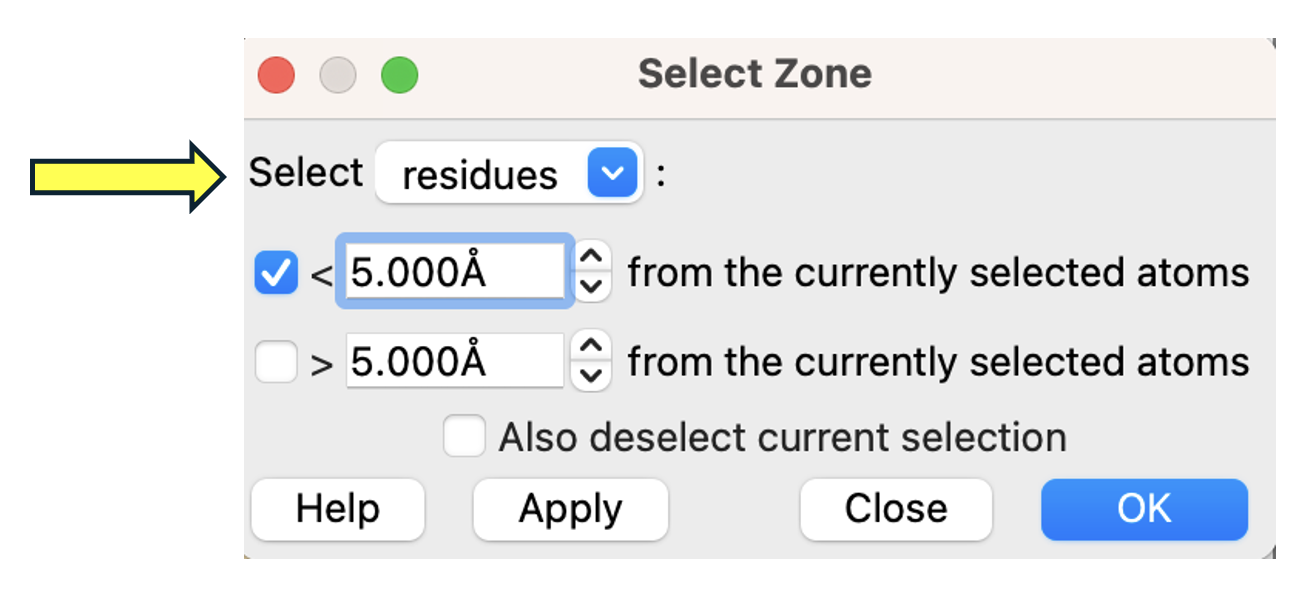
In the popup window, ensure ”Select” is set to “residues” and the “< 5.000Å” box is checked. Click “OK” (Figure 2).
- Then, show these residues as sticks. In the “Home” toolbar selector, use the “Atoms” toolbar and click ”Show.”
- Change the sulfate to spacefill.
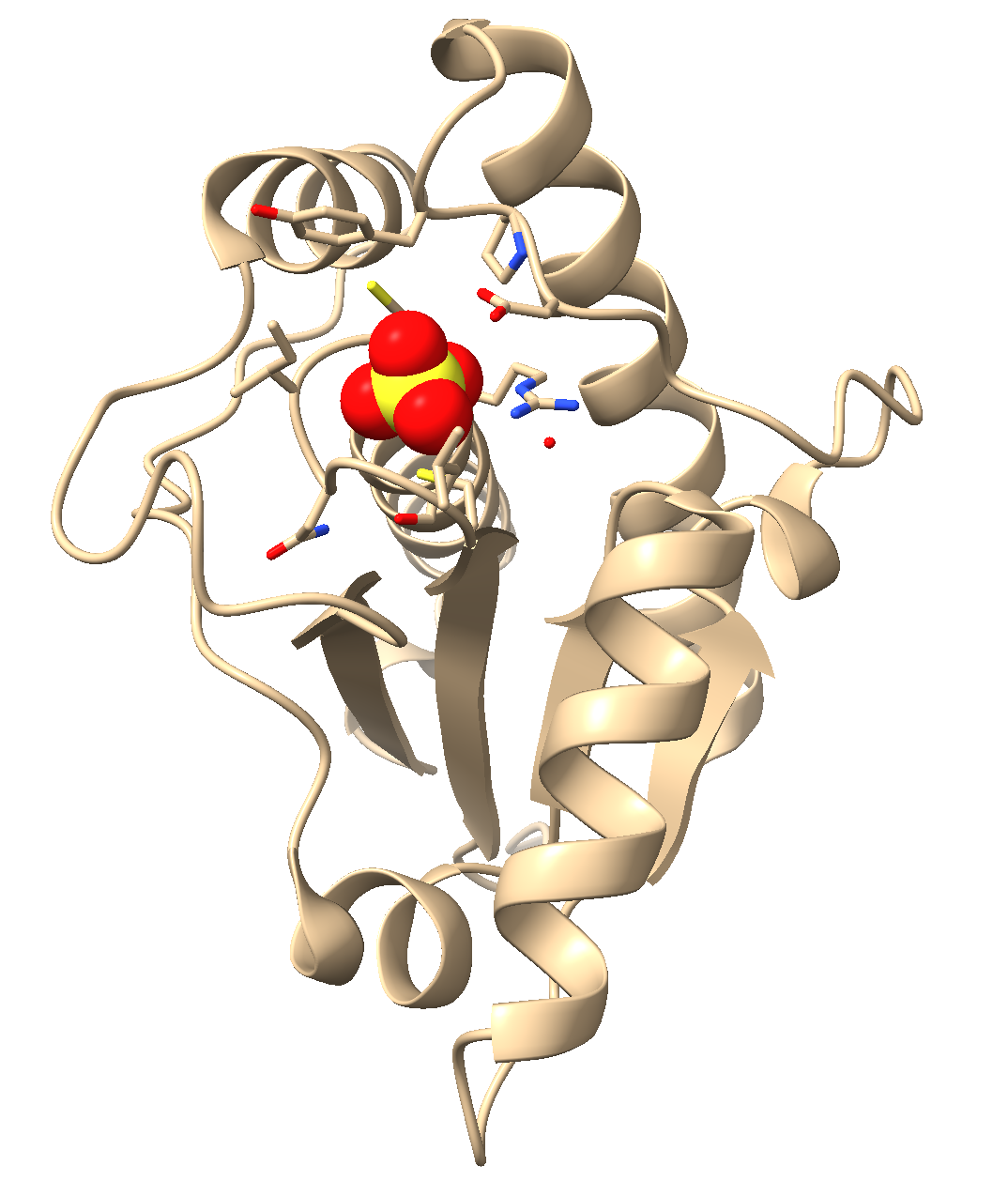
Figure 3: Image of the final activity output
a) In the dropdown menu: Select → Residues → SO4
b) In the “Home” toolbar selector, use the “Styles” toolbar to select “Sphere.”
- Orient the molecule so you can see the sulfate and residues well.
- In the “Home” toolbar selector, using the “Background” toolbar, change the background to white by clicking “White” (if desired).
- (Optional) Save and close the session.
Jump to the next ChimeraX tutorial: VIII.A. ChimeraX Molecular Interactions.
