VIII.C. Mol* Molecular Interactions
Didem Vardar-Ulu and Shane Austin
Overview: This activity demonstrates how to select a macromolecular structure to identify and select nearby objects.
Outcome: The user will be able to focus on a specific part of a macromolecule of interest, display its surrounding residues, and visualize noncovalent interactions within a 5Å radius of the target.
Time to complete: 5 minutes
Modeling Skills
- Focusing on a specific part of the macromolecule of interest
- Selecting groups within 5Å
- Displaying noncovalent interactions
About the Model
PDB ID: 1xww
Protein: Low molecular weight protein tyrosine phosphatase
Activity: hydrolyzes Tyr-OPO32- phosphoester bond
Description: single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
Load the Structure:
- Go to rcsb.org
- In the search bar at the top, type 1xww and press Enter.
- On the landing page, click the “Structure” tab next to the highlighted “Structure Summary” tab to open the user interface and load the structure.
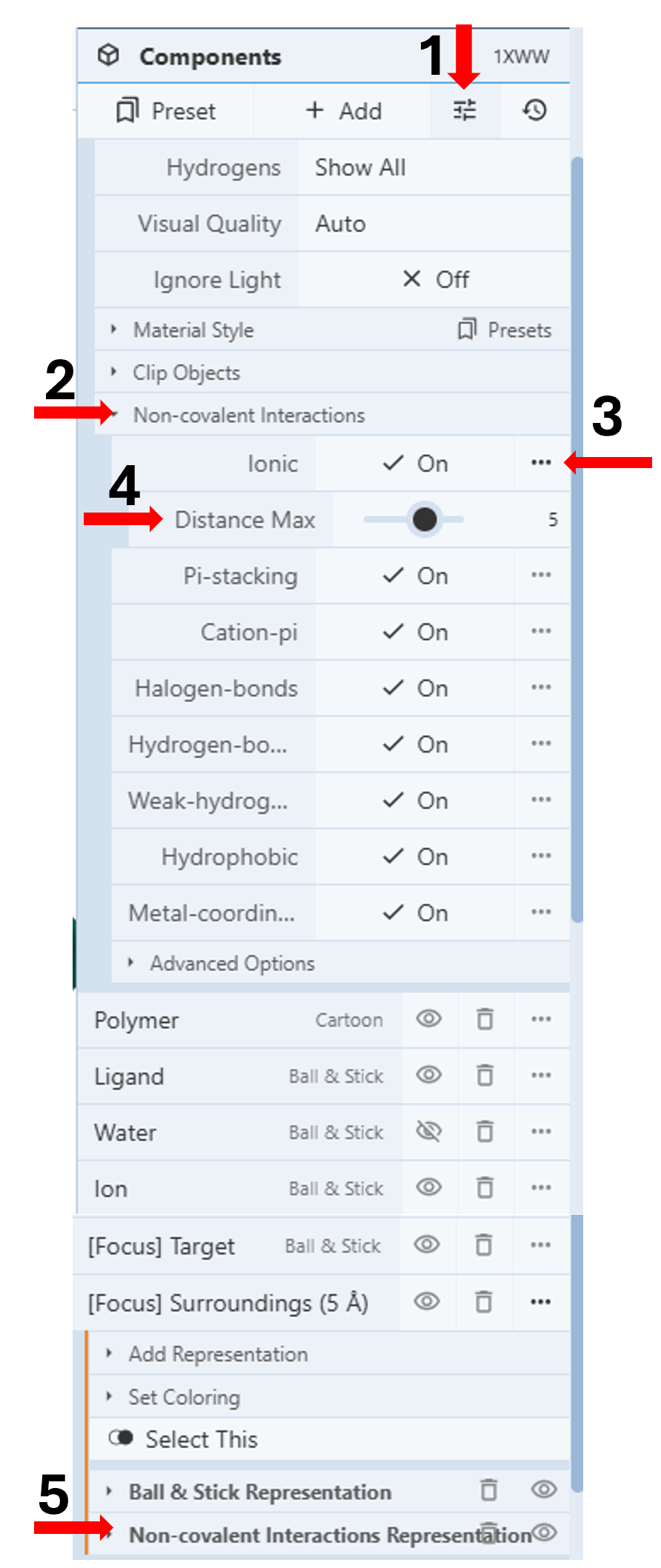
Displaying all non-covalent interactions within 5Å: (Figure 1)
-
-
-
-
-
-
-
-
- To simplify the view, click on the “eye” icon next to the “Water” component listed in the “Structure Panel” to hide it.
- Find the sulfate ion in the 3D canvas or change the displayed sequence to sulfate ion from the component menu.
Note: You can change the color of the sulfate ion using the “Changing Colors” protocol in Chapter II.C. Make sure to Toggle off the “Selection Mode” after changing color.
- Click the sulfate ion to zoom in and display the surrounding groups.
- Click the “Settings” icon in the first row under the Components Panel to access the pull-down menu. (Arrow 1)
- Click “Non-covalent Interactions” (Arrow 2)
- Turn on all the desired interactions by clicking the corresponding on/off toggle.
- Adjust distance cut-offs for each interaction by clicking the three dots next to the corresponding interaction (arrow 3) and then moving the slider to the desired distance. (Arrow 4)
-
-
-
-
-
-
-
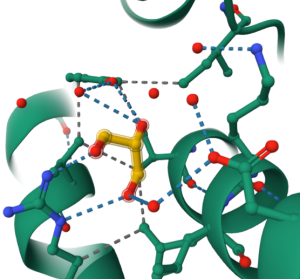
Note: Displayed interactions appear as a separate representation under the temporary [Focus] Surrounding 5Å component. Each interaction is represented by a dashed line in the default color for that interaction. You can modify the attributes for this component by clicking the “Non-covalent Interactions Representation” to access the drop-down menu. (5) You can also make a standalone component for these interactions as described in Chapter VII.C.
Jump to the next Mol* tutorial: IX.C. Manual Measurements
