IX.C. Mol* Manual Measurement
Didem Vardar-Ulu and Shane Austin
Overview: This activity demonstrates how to manually measure distances.
Outcome: The user will be able to measure and label distances between two atoms (e.g., from an atom in a ligand to an atom in the enzyme active site).
Time to complete: 15 minutes
Modeling Skills
- Changing the color of the ligand
- Selecting groups within 5 Å, saving selections (i.e., “components”)
- Manually measure distances between atoms (e.g., from a ligand to an active site amino acid residue)
About the Model
PDB ID: 1eve
Protein: Acetylcholinesterase
Activity:hydrolyzes the neurotransmitter acetylcholine to form acetate and choline
Description: single chain, bound to the drug Aricept (donepezil), covalently bound to five NAG glycan molecules
Steps
Load the Structure:
- Go to rcsb.org
- In the search bar at the top, type 1eve and press Enter.
- On the landing page, click the “Structure” tab next to the highlighted “Structure Summary” tab to open the user interface and load the structure.
Displaying Enzyme active site within 5Å of Aricept (Ligand: 1-benzyl-4-[(5,6-dimethoxy-1-indanon-2-yl)methyl]piperidine):
Color Ligand: (Figure 1)
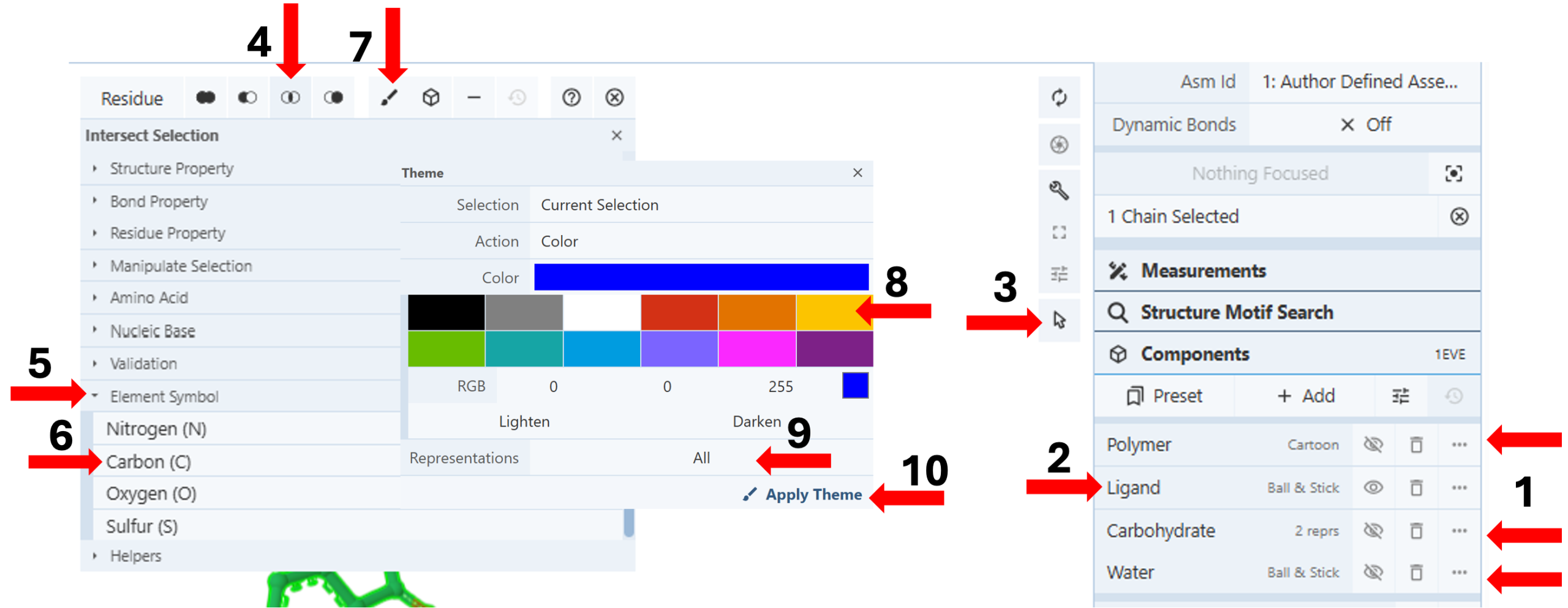
- Hide “Polymer”, “Carbohydrate”, and “Water” components using the “eye” icon in the components panel. (Arrow 1)
- Click “Ligand” on the components menu to focus the drug in the middle of your 3D canvas. (Arrow 2)
Note: You can also change the displayed sequence to 1-benzyl-4-[(5,6-dimethoxy-1-indanon-2-yl)methyl]piperidine from the component menu (see Chapter II.C, Figure 2) and click E20 (the sequence ID for the ligand) in the sequence panel.
- Color the carbons of the ligand in yellow to make it easily distinguishable.
- Activate the “Selection Mode”. (Arrow 3)
- Click the ligand on the 3D canvas to select it.
- Click the “Intersect Selection” icon for a selection menu. (Arrow 4)
- Activate an additional menu by clicking the arrow next to “Element Symbol.” (Arrow 5)
Note: You may need to adjust the scroll-down slider on the right to view the new menu, which opens with individual element names.
- Click “Carbon” to select it. (Arrow 6)
- Click the paint brush icon on the selection bar to open the “Theme” panel. (Arrow 7)
- Choose a desired color (e.g., yellow) using the preset colors (arrow 8) or create a new color using the RGB tool or the Lighten/Darken buttons.
- Click “All” in the cell for representation (9) and choose “Ball & Stick”.
- Reclick the cell for “Representations” (now showing 1 of 16). (Arrow 9)
- Click “Apply Theme”. (Arrow 10)
- Deactivate the “Selection Mode”. (Arrow 3)
- Click the middle of the yellow molecule on the 3D canvas to show all residues and bound waters within 5Å distance from the drug.
- Hide the “Polymer” Component using the “eye” icon next to “Polymer”.
Create a standalone component for the active site within 5Å of the drug without the drug: (Figure 2)
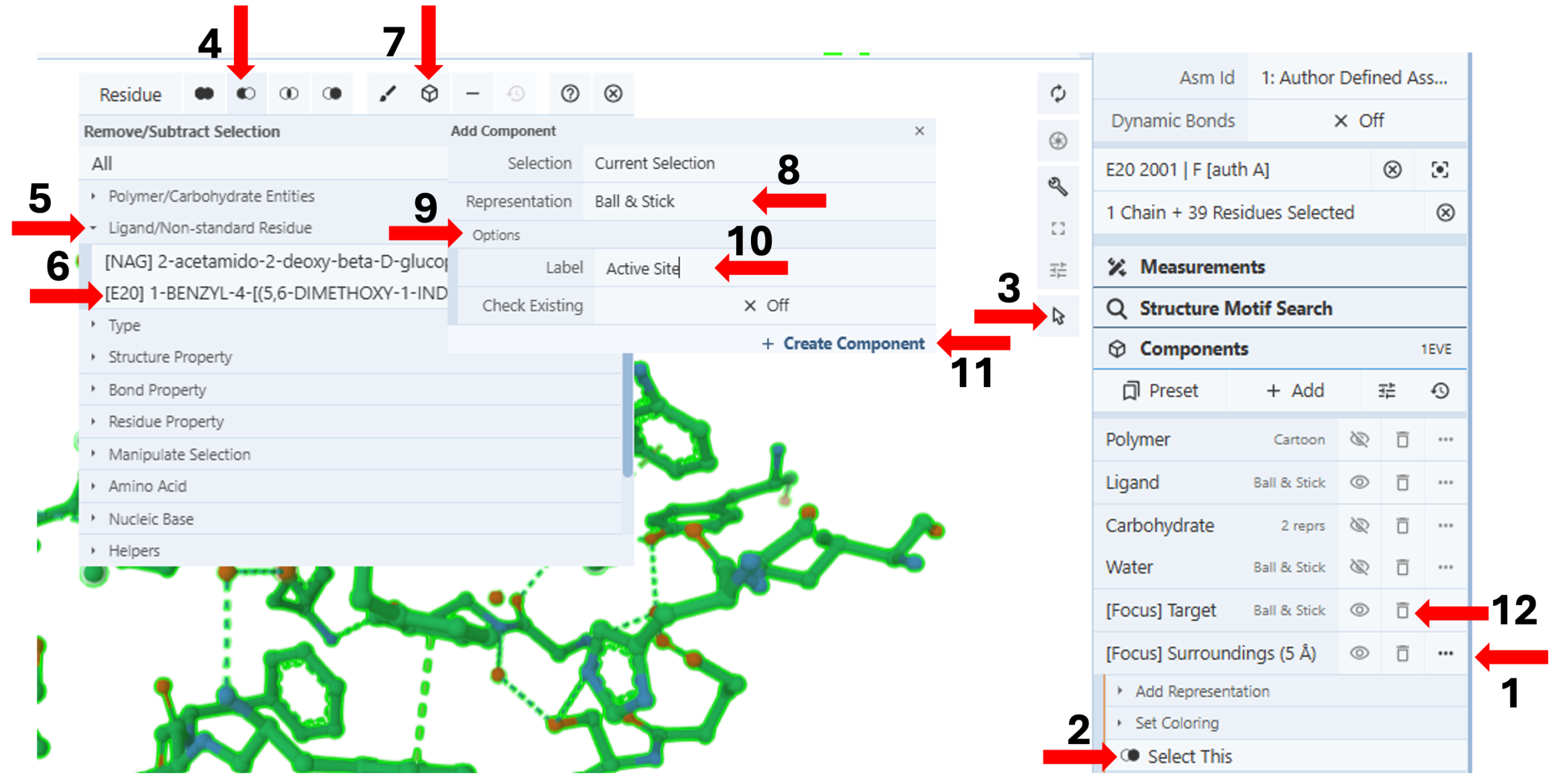
-
- Click the three dots next to “[Focus] Surroundings (5Å)” in the component panel. (Arrow 1)
- Click “Select This”. (Arrow 2)
- Activate the “ Selection Mode”. (Arrow 3)
- Click the “Remove/Subtract” icon for a selection menu. (Arrow 4)
- Activate an additional menu by clicking the arrow next to “Ligand.” (Arrow 5)
- Click “[E20]” to select it. (Arrow 6)
- Click on the “Components Icon” (arrow 7), which will activate a dropdown menu.
- Click on the cell next to “Representation” (default “Create Later”) and choose your desired representation (e.g., Ball & Stick). (Arrow 8)
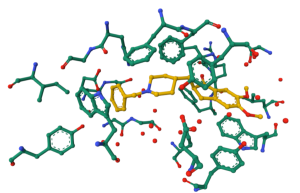
Figure 3: Active site within 5Å of Aricept of 1eve. - Activate the “Options” by clicking on it. (Arrow 9)
- Enter the desired label (e.g., Active Site). (Arrow 10)
- Click “Create Component” (Arrow 11)
- Delete [Focus] Target and [Focus] Surroundings (5Å) temporary components from the components panel using the “trash can” next to the components. (Arrow 12)
Measuring Distances between the Active site and Aricept (Ligand: 1-benzyl-4-[(5,6-dimethoxy-1-indanon-2-yl)methyl]piperidine): (Figure 4)
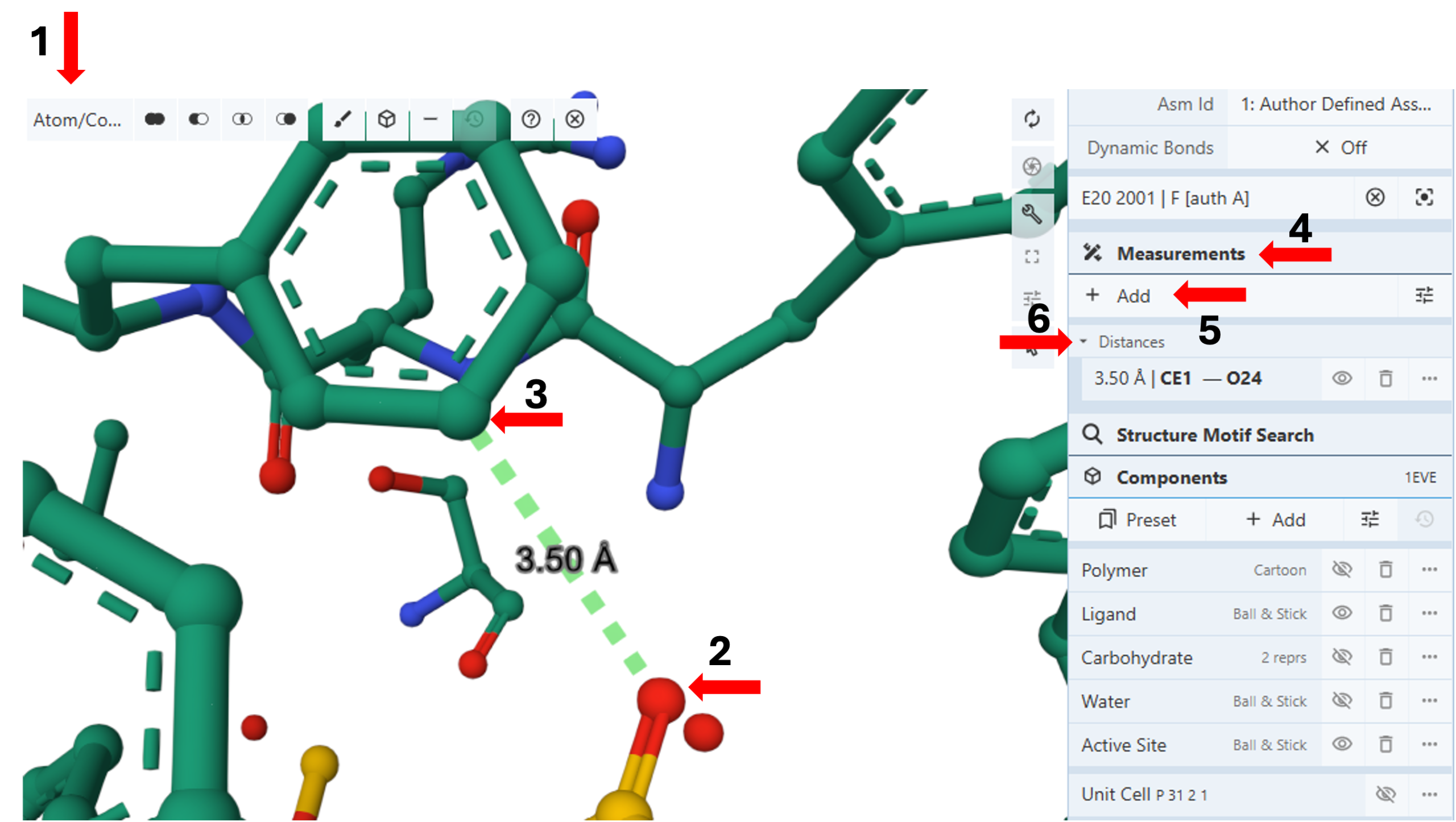
- Change the picking lever of your selection to “Atom/ Coarse element”. (Arrow 1)
- Click the first atom for your distance measurement (e.g., E20, O24) to select it and highlight it in green. (Arrow 2)
- Click the second atom for your distance measurement (e.g., Phe290, CE1) to select it and highlight it in green. (Arrow 3)
Note: Hovering your cursor on any part of the sequence or structure will display information about the PDB ID, model number, instance, chain ID, residue number, and chain name listed at your chosen picking level at the bottom right corner of the 3D canvas.
- Click the “Measurements” in the control panel. (Arrow 4)
- Click “Add”. (Arrow 5)
- Click “Distance”. This will display the distance between the two selected atoms using dashed lines and label it with the distance in angstroms.
Note: This protocol will also make the created distance a standalone entity in the measurement panel. The three dots next to it will allow you to access additional options to modify it.
Jump to the next Mol* tutorial: X.C. Protein-Protein Interface
