VIII.B. iCn3D Molecular Interactions
Henry V. Jakubowski and Kristen Procko
Overview: This activity demonstrates how use one part of a macromolecular structure to select nearby objects.
Outcome: The user will be able to focus on a specific part of a macromolecule of interest, display its surrounding residues, and visualize noncovalent interactions within a 5Å radius of the target.
Time to complete: 15 minutes
Modeling Skills
- Focusing on a specific part of the macromolecule of interest
- Selecting groups within 5Å
- Displaying noncovalent interactions
About the Model
PDB ID: 1xww
Protein: Low molecular weight protein tyrosine phosphatase
Activity: hydrolyzes Tyr-OPO32- phosphoester bond
Description: single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
Load Structure
- Open iCn3D
- In List of PDB, MMDB, or AlphaFold UniProt structures, type 1xww
- Click “load biological unit”.
Select, Save, and Render the Ligand
- Alt+Click on the sulfate ligand in the structure viewer (option+click on Mac).
- Save the selection:
a) In the dropdown menu: Select → Save Selection
b) In the selection popup: type SO4
c) Click “Save”
- Render the ligand in spacefill. Using the dropdown menu: Style → Chemicals → Sphere
Save the Protein as a Selection
- Select the protein chain using the dropdown menus: Select → Select on 3D
- Click on “Chain”, and then alt click the protein (note that you could do this in Analysis menu; see exercise VII.B.)
- Save the selection:
a) In the dropdown menu: Select → Save Selection
b) In the selection popup: type Achain
c) Click “Save”
Note: You could also pick 1XWW_A in Defined sets instead
- Color the protein by secondary structure. In the dropdown menu: Color → Secondary → Sheets in Yellow
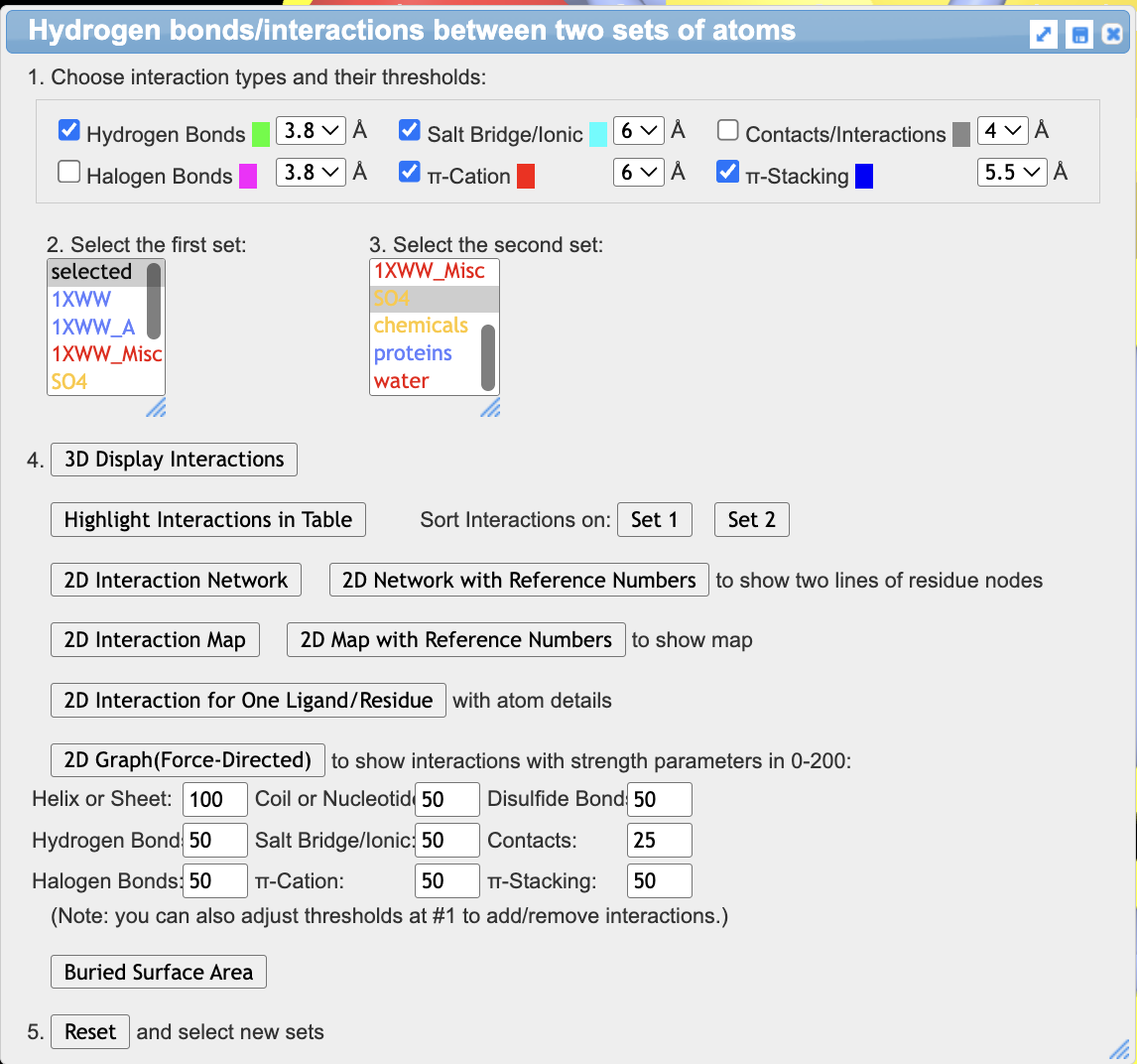
- Show the interactions
a) With the protein still selected, use the dropdown menu: Analysis → Interactions
b) Since the AChain is already selected, as the first set, leave “selected” as the choice.
c) for the second set, click on SO4 in the menu to choose it
d) Check just the noncovalent interactions shown in the image below
e) Click 3D Display Interactions. This will display interacting side chains.
f) Close the window.
- Save the selection of noncovalent interactions.
a) In the dropdown menu: Select → Save Selection
b) In the selection popup: type NCI (for noncovalent interactions)
c) Click “Save”
- Color the amino acids with CPK Coloring.
a) Make the selection. In the dropdown menu: Style → Sidechains → Stick (so that the next step will just color side chains)
b) Apply the color. In the dropdown menu: Color → atom
- Focus on the interactions:
a) In the defined sets popup: Ctrl+click SO4, AChain (1xww_A) and NCI to highlight all
b) In the dropdown menu: View → View Selection (to only see SO4, AChain, NCI)
c) (If needed) Remove the highlight. In the dropdown menu: Select → Toggle Selection
Note: If you closed the defined sets popup, reopen it using the dropdown menu: Select → Defined sets
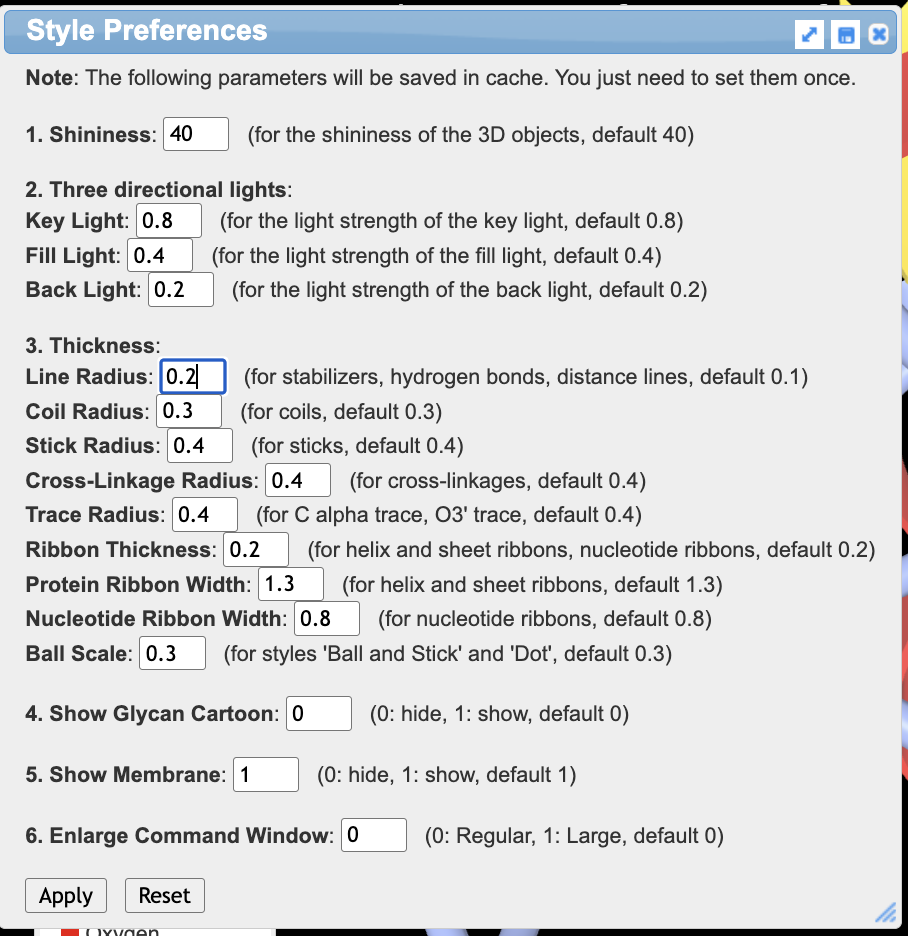
- Set the thickness of the interaction lines.
a) In the dropdown menu: Style → Preferences,
b) In the popup “style preferences” window, adjust: 3. Thickness, Line Radius
c) Type 0.05 in the line radius block
d) Click “apply”
e) Close the popup
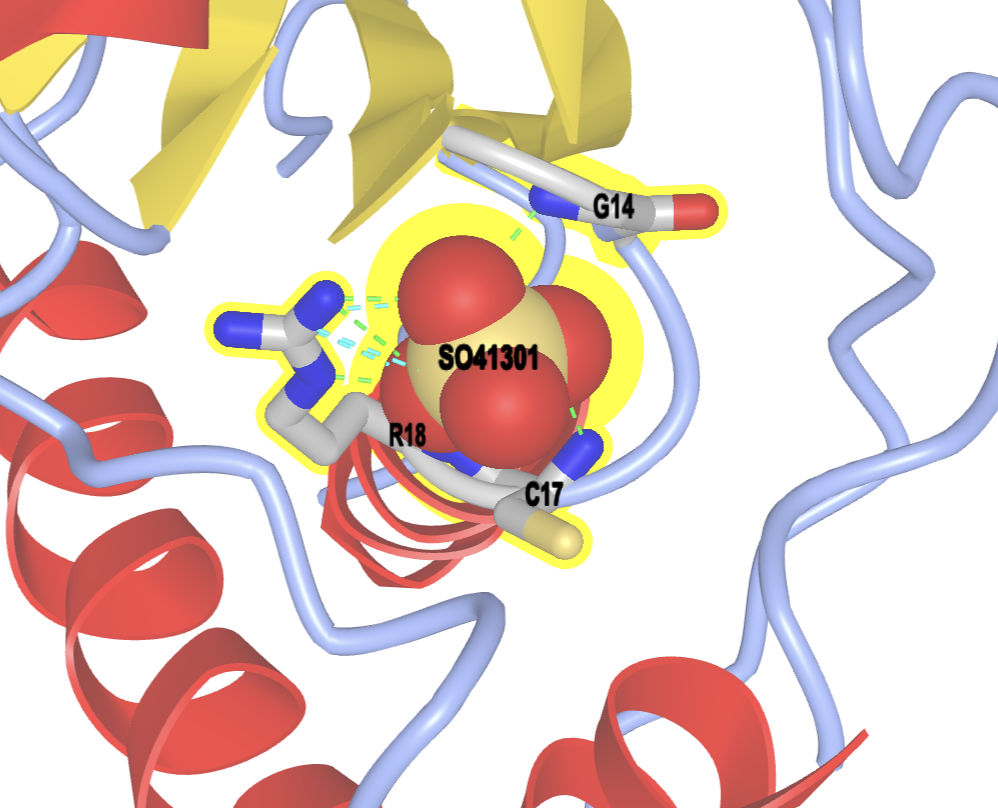
- Label the residues.
a) In the select sets popup, select NCI and SO4
b) In the dropdown menu: Analysis → Label → Per Residue & No
b) Scale the labels. In the dropdown menu: Analysis → Label Scale → pick a number that works for you (try 2.0)
- (Optional) Apply a white background. In the dropdown menu: Style → Background → White
- (Optional ) Save the share link. In the dropdown menu: File → Share Link → copy short link
Jump to the next iCn3D tutorial: IX.B. iCn3D Manual Measurement
