VII.B. iCn3D 5Å Selection
Henry V. Jakubowski and Kristen Procko
Overview: This activity demonstrates how use one part of a macromolecular structure to select nearby objects.
Outcome: The user will be able to select a specific part of a macromolecule of interest, display surrounding residues that are within range to interact with the selection, and save these selections.
Time to complete: 15 minutes
Modeling Skills
- Selecting objects using dropdown menus
- Selecting groups within 5 Angstroms
About the Model
PDB ID: 1xww
Protein: Low molecular weight protein tyrosine phosphatase
Activity: hydrolyzes Tyr-OPO32- phosphoester bond
Description: single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
Load the Structure
- Open iCn3D
- In List of PDB, MMDB, or AlphaFold UniProt structures, type 1xww
- Click “load biological unit”.
Selecting Objects within a Distance
- Alt+Click to select the sulfate in the structure viewer.
Note: Note that you can also select SO4 in the Details tab of the Sequence and Annotations window. See VI.B. iCn3D Selection Tools for details.
- Save the selection. In the dropdown menus: Select → Save Selection. Type SO4 in the box and click save.
- Now, use the SO4 selection to select nearby residues
a) In the dropdown menus: Select → Select by Distance
Note: Our goal is to find all atoms with 5Å from the sulfate. In the next step we will designated 2 sets of species. Set 1 will be the SO4 which is already selected. Set 2 will be what surrounds it. In our case that will be the protein chain (1xww_A) where A stands for the A chain in the PDB file.
b) In the popup window, select the sets. Set 1: click “Selected” (this should already already chosen since the SO4 is still selected);
c) Set Sphere to 5 Angstrom by typing in the block
d) Set 2: click “nonselected” or “1xww_A”
e) Click Display
f) Close the “select a sphere” popup
- Save and render the highlighted groups 5Å from the sulfate
a) Using the dropdown menus: Select → Save Selection
b) For the name, type: 5AfromSO4
c) Render the amino acid residues as sticks. In the dropdown menus: Style → Side chains → Stick
d) Render the water molecule. In the dropdown menus: Style → water → dot
e) Apply CPK coloring. In the dropdown menus: Color > Atom
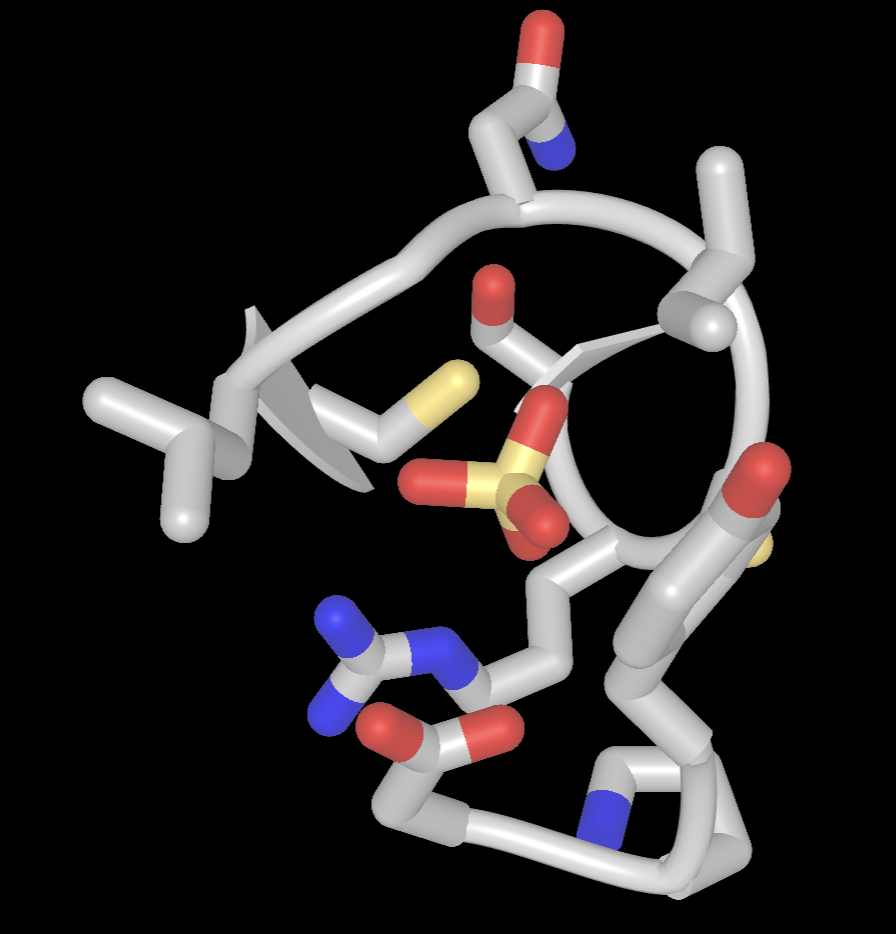
- Limit the view to focus on the sulfate binding site.
a) In the Defined Sets popup, Ctrl+Click on the following selections: SO4, 5AfromSO4
b) In the dropdown menu: View → View Selection
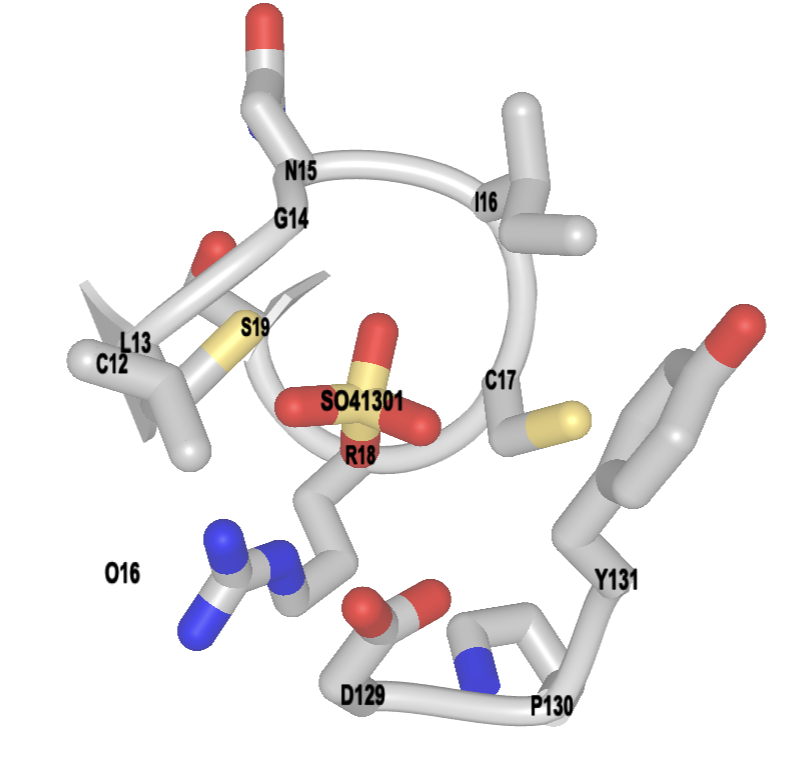
- Label the binding site
a) To label, use the dropdown menu: Analysis → Label → Per Residue and Number
b) To change label color, use the dropdown menu: Analysis → Label → Change Label Color (globally)
c) Pick a color using hexacode, type it in the popup window, click “Display”
c) Resize the label using the dropdown menus: Analysis → Label Scale → pick a number that works for you (try 1.5)
- Change the background color. In the dropdown menus: Style → Background → White
- (Optional) Save the file share link. In the dropdown menu: File → Share Link → copy short link
Jump to the next iCn3D tutorial: VIII.B. iCn3D Molecular Interactions
