5. A Guide to PDB Structures and the RCSB PDB
Brianna Bibel
The Protein Data Bank (PDB)
Structural data for proteins, other macromolecules, and macromolecular complexes is hosted by the Protein Data Bank (PDB), where each structure is given a unique alphanumerical accession code (also called a PDB ID) and a corresponding entry page. The hosted data includes structural models and, for many structures, the underlying experimental data, which typically comes from x-ray crystallography, cryogenic electron microscopy (cryo-EM) or nuclear magnetic resonance (NMR) experiments. In addition to over 230,000 such experimentally-determined (a.k.a. solved) structures as of May 2025, it contains computed structural models (CSMs)–over 1 million as of May 2025.
As an instructor, the PDB is where you can go to find structures to use for classroom activities and assessments. You can download the structures directly (e.g., as PDBx/mmCIF files), then upload to a 3D visualization program (PyMOL, ChimeraX, Mol*, iCn3D, etc., as covered in later chapters). Alternatively, you can fetch the data from said programs by using the 4- or 12-character accession code referred to as the PDB ID.
Note: For the purposes of this OER we will use common terminology, which is not always technically precise. For example, we will use the term structures loosely, e.g., to refer to structural models, which are not true structures but rather models of underlying experimental or computational data. Such terminology might be slightly different from terminology used by structural biologists, however, it is generally accepted and understood.
More than simply a repository, the PDB is a worldwide, community effort to maintain data quality and accessibility. Their vision is to “sustain freely accessible, interoperating Core Archives of structure data and metadata for biological macromolecules as an enduring public good to promote basic and applied research and education across the sciences.” The Worldwide PDB (wwPDB), has centers located on three continents: the Research Collaboratory of Structural Bioinformatics (RCSB) PDB in the United States, the EMBL-EBI’s PDB in Europe (PDBe) and PDB Japan (PDBj). All these institutions hold the same data, but they distribute it in different ways (but still all free). Additional member groups are the Biological Magnetic Resonance Data Bank (BMRB), which hosts NMR data, and the Electron Microscopy Data Bank (EMDB), which hosts cryoEM data. You can find out much more about the wwPDB at their website.
The RCSB PDB
Our team primarily utilizes the Research Collaboratory for Structural Bioinformatics (RCSB) PDB, which is managed by three member institutions of the RCSB: Rutgers; the University of California, San Diego (UCSD); and the University of California at San Francisco (UCSF). This database contains search tools similar to the other PDBs, and also contains a robust collection of educational resources encompassed in a section of the website called PDB 101.
It is easy to search for entries based on their 4- or 12-character accession code, referred to as a PDB ID. Such IDs can often be found at the ends of scientific articles. You can also search the PDB by macromolecule of interest and/or other parameters. For a more detailed guide to searching the PDB, please refer to the next section: “Advanced Search through the RCSB PDB.”
Note: PDB IDs have historically been 4 characters-long; however, due to the tremendous growth of the database, all such possible combinations are expected to be taken by 2029. Therefore, the PDB is transitioning to an extended, 12-character format with the basic structure: pdb_XXXXXXXX
4-Character PDB IDs can be converted to their extended 12-character version by prepending with “pdb_0000.” For example, 9nc3 becomes: pdb_00009nc3. More information can be found at the PDB page “Extended PDB ID With 12 Characters.”
Contents of a PDB Entry
In this section, we will provide an overview of the kind of information you can find on entry pages. When applicable, links to later sections and external resources will be provided for those seeking additional information.
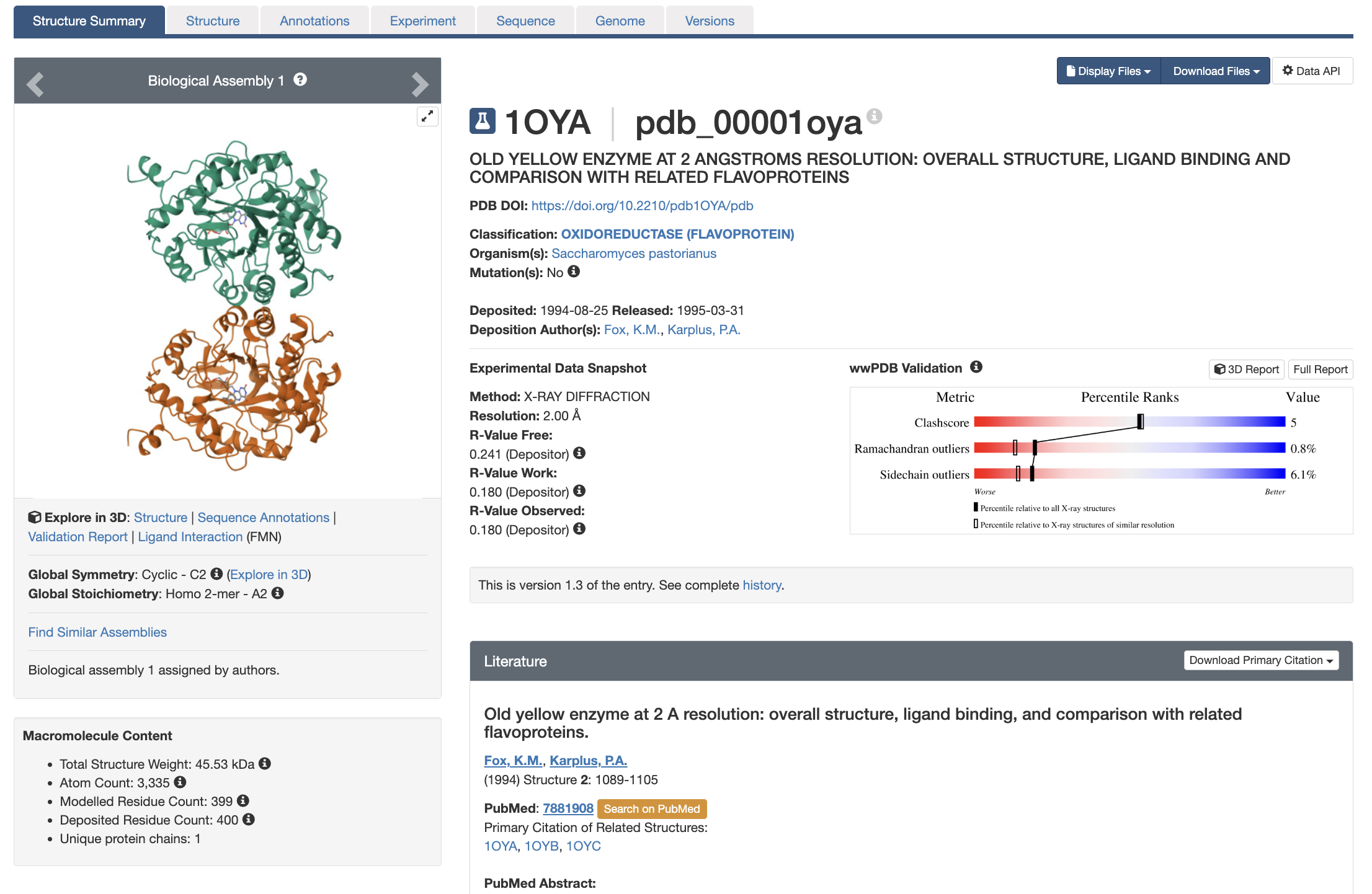
Entries for experimentally-determined structures are indicated by an Erlenmeyer flask icon next to the PDB ID and CSM entries are indicated by a computer icon. The main page of an entry is the Structure Summary page (example shown in Figure 1). It has multiple components that allow you to quickly see:
- Quality of the structural model and underlying experimental data (A guide to evaluating quality can be found in this book, section Evaluating Structure Quality)
- What’s in the structure (both macromolecules and small molecules)
- Experimental details
- Corresponding publications, when applicable
There are also drop-down menus that allow you to select files to display and/or download.
Structural Files
The PDB provides options to download the data in several different formats (selected from the dropdown “Download Files” menu at the top right of a Structure Summary page). The standard format is PDBx/mmCIF (Protein Data Bank Exchange macromolecular Crystallographic Information File), whose files end in .cif. This file format is more flexible than the more limited legacy PDB format, whose files end in .pdb. A detailed guide to the syntax of PDBx/mmCIF is freely available for readers interested in learning more.
For multiple reasons, including incompatibility of PDB files with larger structures and extended PDB IDs, the PDB recommends that everyone use CIF files exclusively. More information on these file types can be found at the PDB Page: File Formats and the PDB.
Annotated structure file examples
Typically, users will wish to select the “Biological Assembly” from the drop-down menu. More information about biological assemblies can be found later in this chapter.
Note: The “Display Files” dropdown menu, located next to the “Download Files” menu at the top right of a Structure Summary page allows you to view structure files (mmCIF and/or legacy PDB) in a separate browser tab. Selecting an option with “(Header)” in the title will display only the header sections of the files.
Features of the Structure Summary Page
Basic Information
Just below the PDB ID and structure title on the Structure Summary page (Figure 1), you will find several key pieces of information, including the DOI of the associated journal article (if available), deposition date and authors, and organism. In cases where the protein and/or nucleic acid were recombinantly expressed, corresponding information will be displayed in an “Expression System” line.
After this information is an important line to check: Mutation(s). This line will have either a Yes (indicating the macromolecular structure has a sequence that is different from the reference, “wild-type,” sequence) or No (indicating the macromolecular structure has a sequence is identical to the reference sequence). Depending on course level, instructors may wish to select (or have students select) only structures without mutations when possible. In cases where structures containing mutations are used, instructors should be prepared to discuss with students why the residues they observe in the structural models are different from those expected from the wild-type sequence.
The Experimental Data Snapshot section includes information about data and model quality, as explained in the next chapter: Evaluating Structure Quality.
When available, information about a corresponding publication is provided under Literature.
Macromolecules Section
The next page section is Macromolecules (Figure 2, Top). Here, you will find listed each protein and nucleic acid molecule contained in the structure, each of which will be designated as a “Chain” and provided an accompanying alphabetic chain identifier. If there is more than one copy of such a molecule in the structure (e.g. as is the case with homomultimers), each copy will be provided a separate chain identifier. In this section, you will see a quick overview of each macromolecule including size, organism it comes from, any introduced mutations, and information about the corresponding protein (including a link to UniProt, a website for protein sequence and functional information), when applicable.
Below that, the Sequence Annotation section provides further information about the macromolecule sequence. One of the most valuable things to check here is the lane entitled “Unmodeled”. Gray shading indicates which, if any, regions of the macromolecular chain are not present in the structural model, despite being physically present. Such unmodeled regions are often due to movement and/or heterogeneity in the underlying macromolecules–this prevents them from being visible in the experimental data. Hence, such regions are left out of the model; when viewed in a modeling program, they may be indicated by grayscaling of the sequence, and/or dotted or dashed lines in the structural model. This is a frequent area of confusion for students, and more information about it can be found in the PDB 101 article Missing Coordinates and Biological Assemblies.
Below Macromolecules is a Small Molecules section (Figure 2, Bottom), which provides information about other molecules and ions present in the structure. Such molecules may be physiologically relevant (e.g. enzymatic substrates, regulatory ligands, metal cofactors) or they may be experimental artifacts (e.g. glycerol from the sample cryoprotectant). Similarly to the macromolecules, each small molecule receives a designated chain identifier.
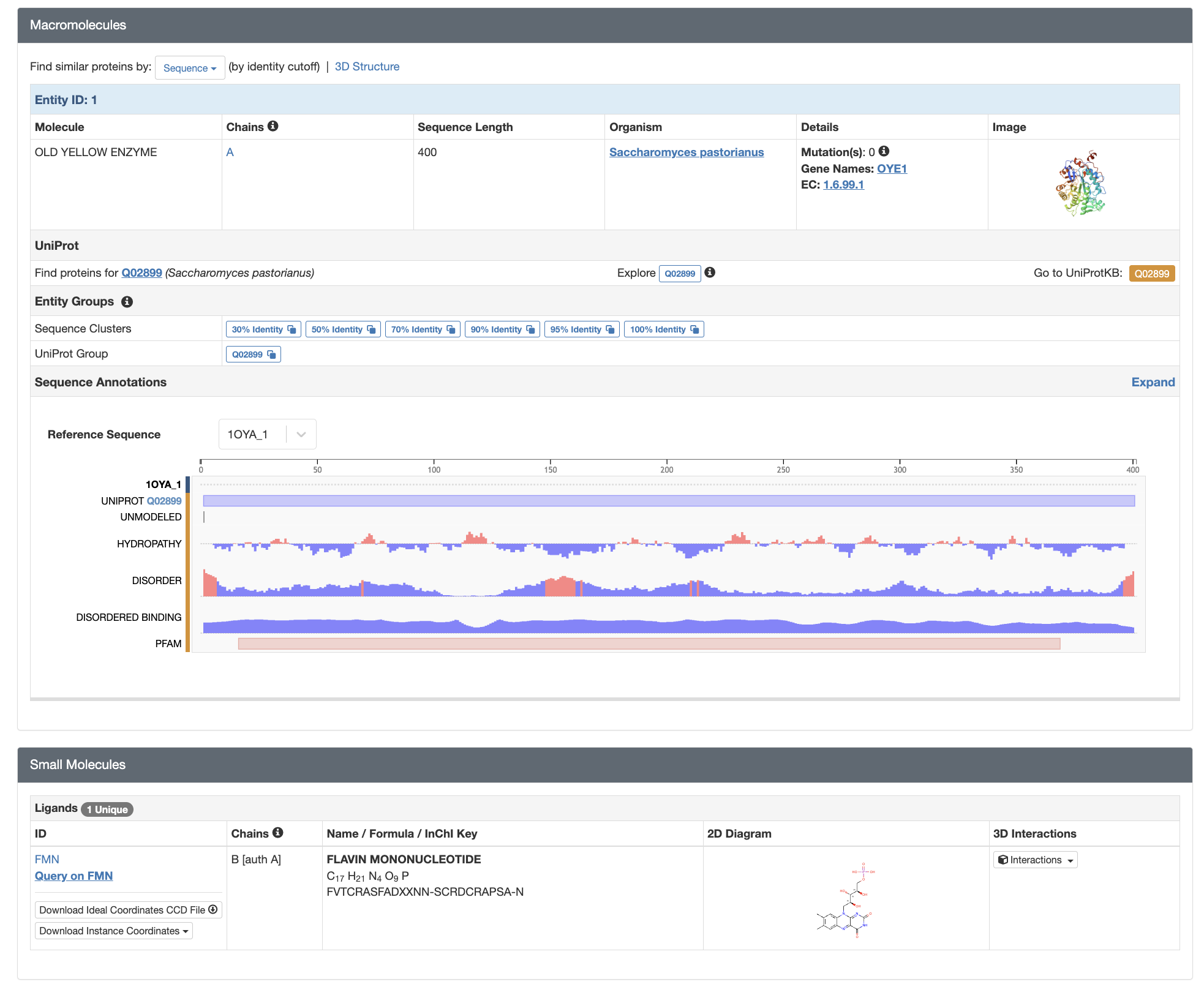
Finally, the Structure Summary page includes overviews of Experimental Data and Validation” and “Entry History.”
Additional Sections of an RCSB PDB Entry
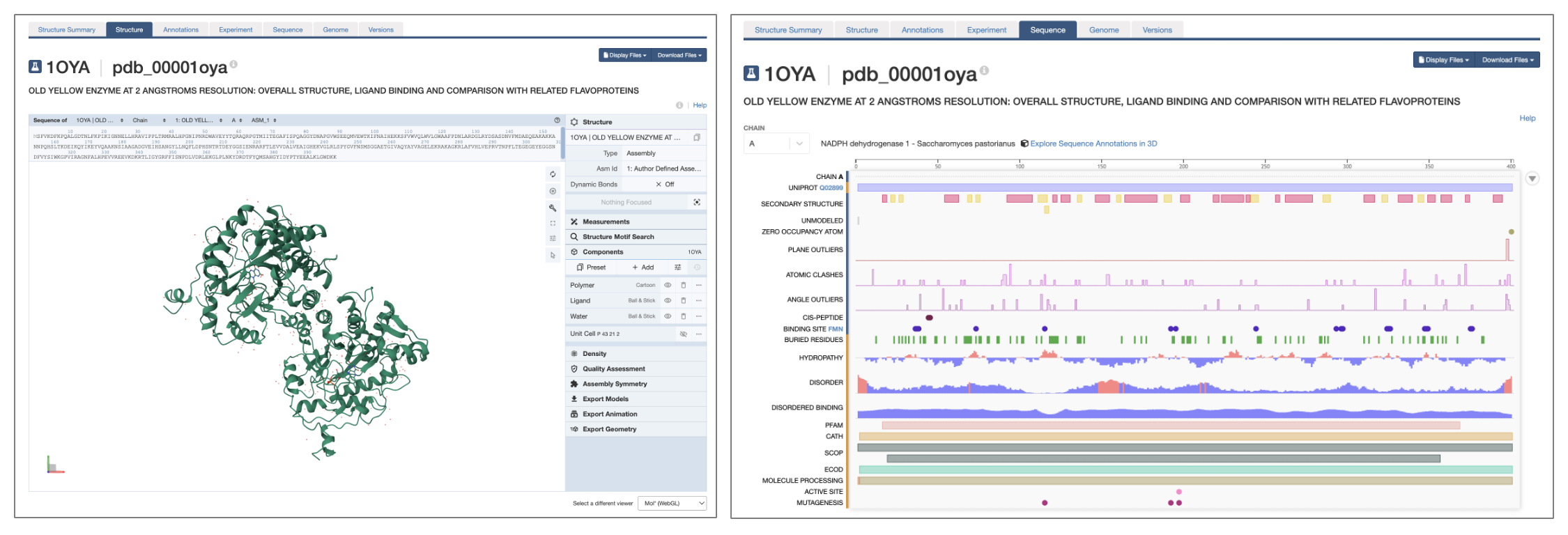
Next to the tab for the Structure Summary page, you will find tabs for a number of other pages with more in-depth information (Figure 1). Depending on the type of entry, these tabs may include:
- Structure (Figure 3, left)
- An integrated 3D molecular viewer that, by default, uses Mol*, but also provides a JMol option. Detailed information about working with Mol* can be found in I.C. Getting Started with Mol* (Molstar) and associated chapters.
- Annotations
- Information about domains and motifs contained in the structure, protein families, etc.
- Experiment (only for experimentally-determined structures)
- Detailed data collection and processing information
- Sequence (Figure 3, right)
- Annotation tracks mapping structural properties (e.g. secondary structure), biophysical properties (e.g. hydrophobicity, disorder), model properties (e.g. clashes, outliers), and functional properties (e.g. active sites, binding sites) to the sequences of the chain(s)
- Genomes
- Where in the genome corresponding proteins and/or nucleic acids are encoded
- Versions
- History of the PDB entry
A Note About Biological Assemblies and Asymmetric Units
When using the built-in Structure viewer in the RCSB PDB, you will be shown the biological assembly (aka biological unit) of a protein or complex by default. This is the form of the molecule that is thought to be the functional unit in an organism and is typically the form you are interested in working with.
To download the biologically relevant .cif file from the PDB, select “Biological Assembly” from the dropdown menu on the Structure Summary page. In the case that multiple biological assemblies are provided, we recommend users examine the different assemblies in the Structure viewer to select the most relevant structure for their purposes.
Sometimes, if you look at the model from a X-ray crystallography structure in the 3D viewer, you will see something different than expected–for example, you may see multiple copies of a monomeric protein, or a single monomer of an homomultimeric protein. This is because in these cases, rather than the biological assembly, you are being shown the asymmetric unit: the smallest repeating part of the crystal from which the structure was determined. If you know the structure of the asymmetric unit and the dimensions of something called the unit cell, you can recreate the whole crystal just using symmetry operations (rotate, move left, move up, etc.)
The asymmetric unit might itself contain more than one copy of the biological assembly (e.g., with one upside down and the other right-side up). The copies of the biological assembly within an asymmetric unit are identical in their sequence, but they might be slightly different in their conformation, and during the modeling they are therefore treated separately. In the PDB, these “different copies” within the asymmetric unit are called “instances.” The PDB has a great guide for anyone interested in learning more: Introduction to Biological Assemblies and the PDB Archive
When using the built-in Structure viewer in the RCSB PDB, you can toggle back and forth between these different forms. In the upper right corner under Structure, click to the right of the word “Type” to reveal a menu (Figure 4). The biological assembly can be found under “Assembly,” which is the default (Figure 4, left) and the asymmetric unit can be displayed by clicking “Model” (Figure 4, right).
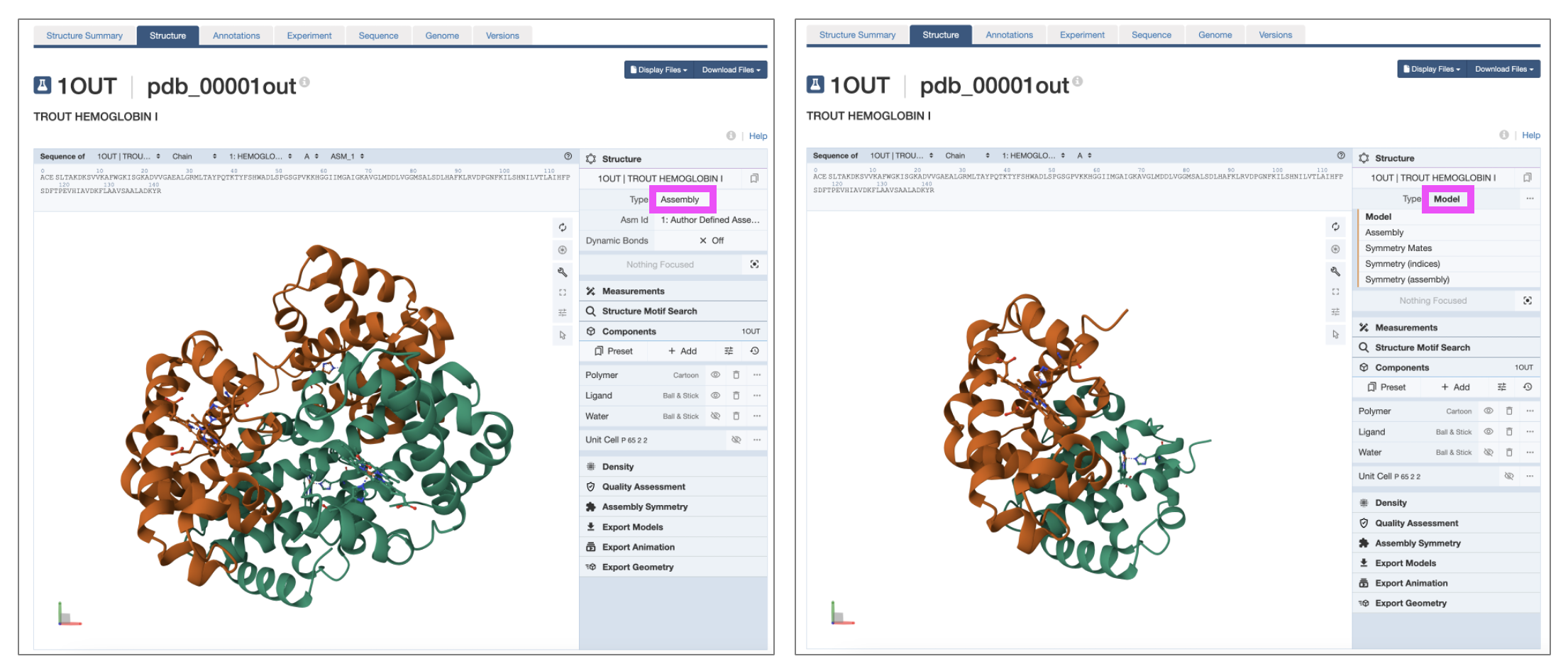
Note: The biological assembly is not always reported and/or identified and therefore the displayed “Assembly” structure may not always be the biologically-relevant form.
