2. Essential Resources for Pre-Rendered Structures
James Nolan and Josh Beckham
Introduction
Showing students how to manipulate structure models generates student interest in learning to do it themselves, by showing them what is possible and how it is useful. Fortunately, students can be guided through a number of pre-rendered structures generated by others, to tour important macromolecules. Here, we describe some interactive resources that we use in our teaching when we don’t have time for students to render a structure themselves.
The resources featured on this page are:
- Proteopedia
- The Online Macromolecular Museum Exhibits
- Fundamentals of Biochemistry (Jakubowski and Flatt online text)
- Molviz Atlas of Macromolecules
- Online Biomolecular Visualization Tools (iCn3D, FirstGlance, and Mol* at PDB)
Proteopedia
Proteopedia (Hodis, 2008; Prilusky, 2011) is a crowd-sourced repository of biomolecular structures presented through wiki web pages. The style of the pages varies along with their authors. A typical page displays a 3D structure in a window alongside a descriptive text. A main strength of Proteopedia as a teaching tool lies in the ability to embed links to scripts that render the structure to best illustrate the points described in the text. The reader can manipulate the model on a guided exploration without learning how to use the rendering program, JSmol, which powers the pages. Figure 1 shows an example: the page opens with an applet that renders the structure as wireframe colored in CPK colors.
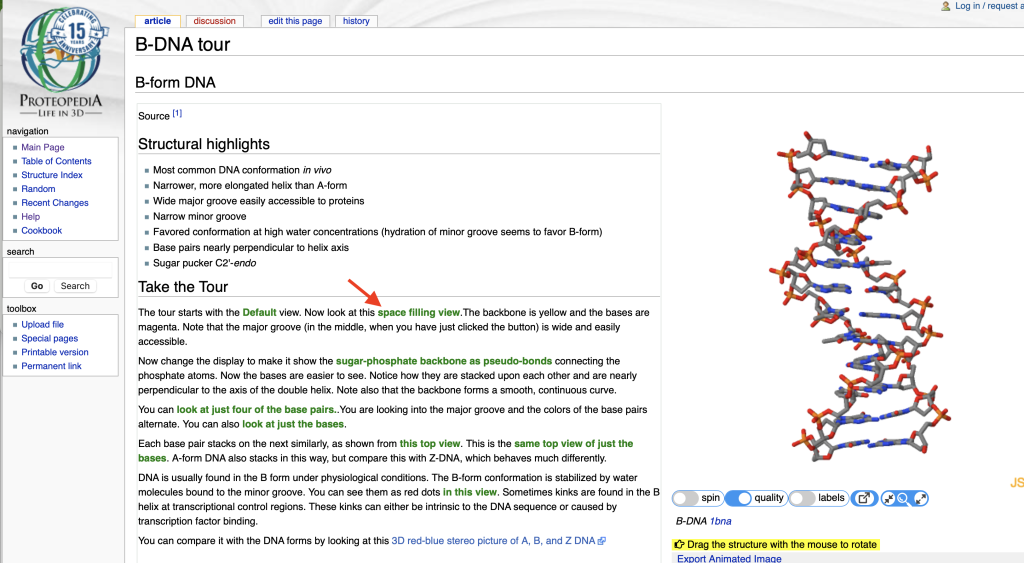
Clicking on the green link indicated with the red arrow causes the applet to show the rendering seen in Figure 2 to guide the reader:
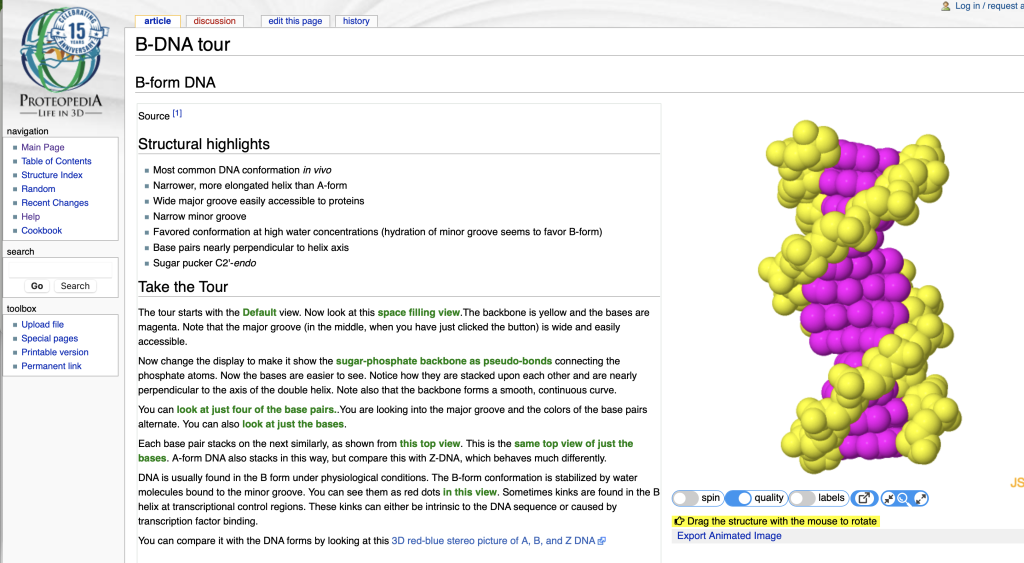
In this case, the new view shows the sugar phosphate backbone colored yellow and the bases are in magenta; all atoms are shown in spacefill for visual clarity. In both views, the structure rendering window can be manipulated by the viewer to explore the structure model. If the user has trouble reorienting the view, they can click on the provided green links to get back to the original orientation. The interactive model window has buttons below it to toggle Start/Stop of model spinning, Low/Higher resolution rendering, On/Off labels, popping the window out of the page, and adjusting the size of the window on the page. You can try out the page shown above at B-DNA tour on Proteopedia.
Note: Some Proteopedia pages open with displays of simple animated GIFs, but have a “Display Interactive Model” button to launch the JSmol guided interactive display of the structure.
Two downsides of Proteopedia are the variability of content and the relatively low-resolution renderings of macromolecules (even in high-quality mode).
Examples of Proteopedia Page Collections Used in Teaching
Proteopedia’s ease of use for readers of all levels of experience has spawned its extensive use as a teaching tool. With over 10,000 entries generated by over 4,000 registered users, finding the best fit can be a challenge. Fortunately, there are several pages with collections of Proteopedia pages that are suitable for Biochemistry courses.
Proteopedia:Biochemistry in 3D
Proteopedia:Biochemistry in 3D lists Proteopedia pages that can serve as supplements throughout a typical one-semester Biochemistry course. Topics include:
- Protein structure: Introduction_to_protein_structure, Alpha helix (includes a quiz at the end), Tutorial:Ramachandran_principle_and_phi_psi_angles, and many more
- Protein function: Ann Taylor’s hemoglobin along with many others on hemoglobin
- Enzyme activity and mechanism: Chymotrypsin, Lysozyme, Hexokinase, Citrate Synthase (shows movement of protons and electrons in reaction mechanism)
- Signaling: Insulin receptor (extensive discussion of structure and conformational changes)
- Carbohydrates: Hexoses, Polysaccharides
- Genes to Proteins: DNA, RNA, tRNA, DNA polymerase (overview page with some structures plus links to additional pages on different versions of DNA polymerases), Ribosome (not a guided tour, but many scripts to toggle display of different components on and off to help exploration)
Teaching Scenes, Tutorials, and Educators’ Pages
Teaching Scenes, Tutorials, and Educators’ Pages includes links to numerous Proteopedia pages appropriate for student use, some of which are:
- Acetylcholinesterase: Substrate traffic and inhibition, Mary G. Acheampong, et al
- Ricin, Ann Taylor
- Serine Proteases , Amy Kerzmann
The list is organized by concepts and it also lists other resources, such as syllabi and quizzes on Proteopedia.
Interactive 3D Complements for Journal Articles (I3DC)
Another collection of existing proteopedia pages is the Proteopedia:I3DC (Interactive 3D Compliments to journal articles). These pages are composed by journal article authors to act as supplemental materials for their 3D structure papers. Articles from numerous traditional research journals, including Cell, Journal of Molecular Biology, Protein Science, and Science can be found. These could be used in the classroom for Journal Clubs or for Case Studies.
Of particular interest to educators is the selection of articles from Biochemistry and Molecular Biology Education (BAMBED): Category:Featured_in_BAMBED. Example pages include:
- Eukaryotic Protein Kinase Catalytic Domain, Alice C. Harmon
- A practical guide to teaching with Proteopedia, Claudia Castro, et al. (Castro, 2021) is a collection of Proteopedia activities used by the authors in their Biochemistry courses. Links provided include a list of structure models in an interactive supplement to Figure 1 of their article, plus supplementary materials for activities for students to expand their understanding of molecular structures. Example learning outcomes are listed for the activities used by the authors.
Help:Teaching with Proteopedia
Help:Teaching with Proteopedia gives instructors an overview of options for using Proteopedia in your course, from finding suitable existing pages to authoring your own pages and creating student accounts. Users do not need to have an account to view/interact with existing structure pages, only to edit them.
BioMolViz Proteopedia page
The BioMolViz Proteopedia page shows Proteopedia examples of the BioMolViz Framework (Dries, 2017) of themes for Biomolecular visualization learning goals. Instructors can select which Framework themes are most appropriate for theirstudents as they progress in their understanding of molecular structure.
The Next Step: Making Your Own Proteopedia Pages
For more advanced students and instructors, users can request an account to compose pages of their own. “Sandbox” pages can be used while learning to construct and edit pages. Using the word “Sandbox” in the page name indicates that the page should not be edited by other users while under construction; sandbox pages can also be edited while using a generic student account, so that students do not need individual accounts to compose assignments. Sandbox pages can also be protected to ensure that only the author can edit them. The Proteopedia:Primer page gets new users started on setting up a new page and the Proteopedia:DIY:Scenes shows them how to compose scenes of structures on their page using the Scene Authoring Tool to select and manipulate specific components of their structure. There is also a Proteopedia:Video_Guide page to help beginners to get started with authoring.
The Online Macromolecular Museum Exhibits
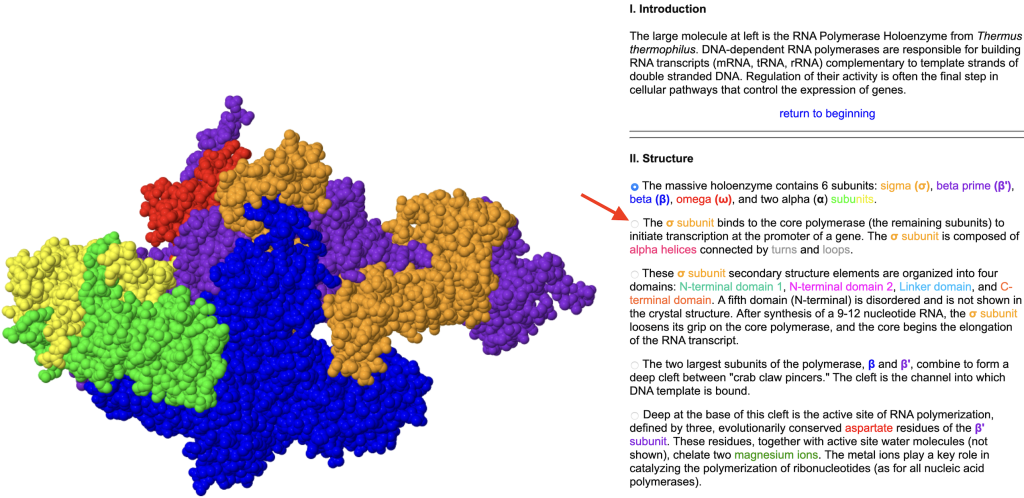
The Online Macromolecular Museum (OMM) has curated tutorials similar to Proteopedia. The collection is not crowd-sourced and is smaller, but still covers many important topics to supplement instruction in Biochemistry. Like Proteopedia, OMM uses JSmol for rendering structures; the scripts that manipulate the display are activated by clicking small buttons in the text.
Approximately 60 entries available to view, examples include:
- An Introduction to DNA Structure
- RNA Polymerase (Figure 3 below)
- An Introduction to Ribosome Structure
Fundamentals of Biochemistry (Jakubowski and Flatt online text)
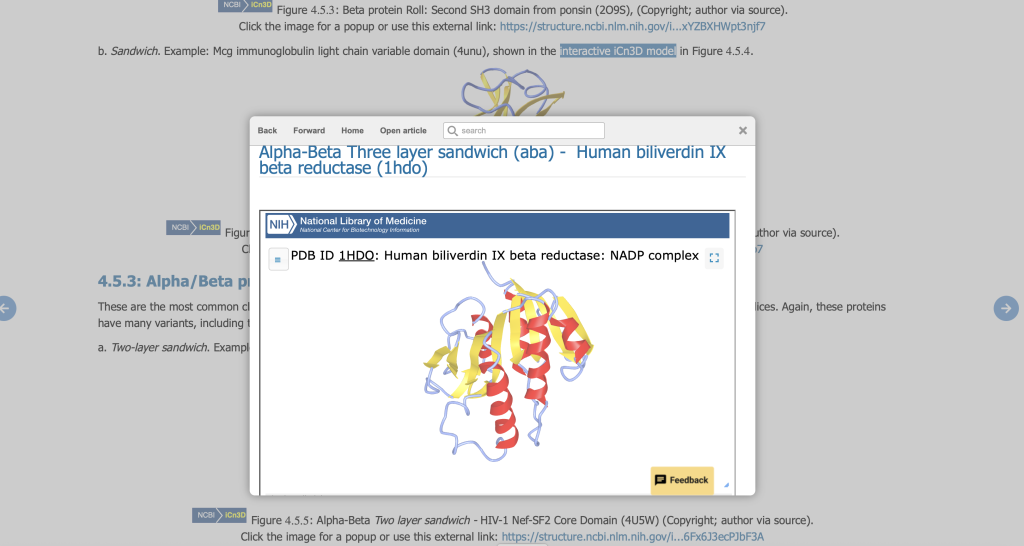
Fundamentals of Biochemistry, by Henry Jakubowski and Patricia Flatt (Jakubowski, 2022), is an excellent example of how the web-based iCN3D rendering program can be used as an educational tool in an online Biochemistry text. Throughout the text, structure figures include links to live, interactive versions in iCN3D popup windows as shown in Figure 4. The windows are small, so the reader can manipulate the structure while still reading the chapter, but can also be resized to fullscreen for a closer look. Chapters 4 through 8 are full of informative examples.
Here is a link to the categorized iCn3D interactive models embedded in the text. The models in these lists don’t contain textual explanations (as of 5/25). To access the actual iCn3D model in the text with detailed explanations, visit the Fundamentals of Biochemistry search page and enter in the PDB or AlphaFold ID shown in the list page. Search the Fundamentals of Biochemistry page for the same ID to find the iCn3D model and textual explanations.
Molviz Atlas of Macromolecules
The Molviz Atlas of Macromolecules is a collection of links to structure renderings for a wide range of molecules of interest: enzymes, other proteins, nucleic acids, and viruses. The links provided lead to Proteopedia pages which seem to be generated automatically based upon the PDB accession number. Alternative links lead to FirstGlance renderings, which are also automatically generated Proteopedia pages based on PDB accession number, but with a different format.
Online Biomolecular Visualization Tools
iCN3D
ICN3D (Wang, 2020; Wang, 2022) is a web-based structure rendering program that is relatively easy to learn (see section I.B and sections in following chapters to get started using the program and making your own figures). If you are not ready to set up your own figures, you can find premade, live figures created by other authors, such as those found in the iCN3D gallery on the iCN3D introduction page.
Our chapters in this book iCn3D: Program Version, Download, & Additional Resources and I.B. Getting Started with iCn3D will help you get up to speed to interact with structures.
FirstGlance
FirstGlance.jmol.org is a good starting point if you know a PDB structure that you are interested in, but do not want to spend time mastering a new program to render it. Just enter the PDB accession code and a web page is generated with easy-to-find tools to study the structure.
Mol* at PDB
In a similar vein to FirstGlance, you can get started interacting with the structure you have found at the PDB (www.rcsb.org) just by clicking on the Structure tab for an entry. Our chapters in this book Mol* (Molstar): Program Version, Download, & Additional Resources & I.C. Getting Started with Mol* (Molstar) will help you get up to speed to interact with the structure. You can also find help at the RCSB PDB website.
References
Castro, C., Johnson, R. J., Kieffer, B., Means, J. A., Taylor, A., Telford, J., … & Theis, K. (2021). A practical guide to teaching with Proteopedia. Biochemistry and molecular biology education, 49(5), 707-719. https://doi.org/10.1002/bmb.21548
Dries, D. R., Dean, D. M., Listenberger, L. L., Novak, W. R., Franzen, M. A., & Craig, P. A. (2017). An expanded framework for biomolecular visualization in the classroom: Learning goals and competencies. Biochemistry and Molecular Biology Education, 45(1), 69-75. https://doi.org/10.1002/bmb.20991
Hodis, E., Prilusky, J., Martz, E., Silman, I., Moult, J., & Sussman, J. L. (2008). Proteopedia-a scientific’wiki’bridging the rift between three-dimensional structure and function of biomacromolecules. Genome biology, 9, 1-10. http://dx.doi.org/10.1186/gb-2008-9-8-r121
Jakubowski, H., Flatt, P., Agnew, H., & Larsen, D. (2022). Fundamentals of biochemistry, a free and new LibreText book for undergraduate courses. The FASEB Journal, 36. https://doi.org/10.1096/fasebj.2022.36.S1.R4590
Prilusky, J., Hodis, E., Canner, D., Decatur, W. A., Oberholser, K., Martz, E., … & Sussman, J. L. (2011). Proteopedia: a status report on the collaborative, 3D web-encyclopedia of proteins and other biomolecules. Journal of structural biology, 175(2), 244-252. http://dx.doi.org/10.1016/j.jsb.2011.04.011
Wang, J., Youkharibache, P., Zhang, D., Lanczycki, C. J., Geer, R. C., Madej, T., … & Marchler-Bauer, A. (2020). iCn3D, a web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics, 36(1), 131-135. https://doi.org/10.1093/bioinformatics/btz502
Wang, J., Youkharibache, P., Marchler-Bauer, A., Lanczycki, C., Zhang, D., Lu, S., … & Ge, Y. (2022). iCn3D: from web-based 3D viewer to structural analysis tool in batch mode. Frontiers in molecular biosciences, 9, 831740. https://doi.org/10.3389/fmolb.2022.831740
