XI.B. iCn3D Superimposition
Henry V. Jakubowski and Kristen Procko
Overview: This activity demonstrates how to align two objects in the modeling program.
Outcome: The user will be able to superimpose two similar macromolecules to examine commonalities and differences
Time to complete: 5 minutes
Modeling Skills
- Superimposing objects
About the Models
PDB ID: 1xww & 5pnt (different isozymes)
Protein: Low molecular weight protein tyrosine phosphatase
Activity: hydrolyzes Tyr-OPO32- phosphoester bond
Description: single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
- Open iCn3D
- Open and align the two structures. In the dropdown menu: File → Align → Protein Complexes → Two PDB Structures
Note: The menus will have different options depending on whether you have selected “simple menus” or “all menus”.
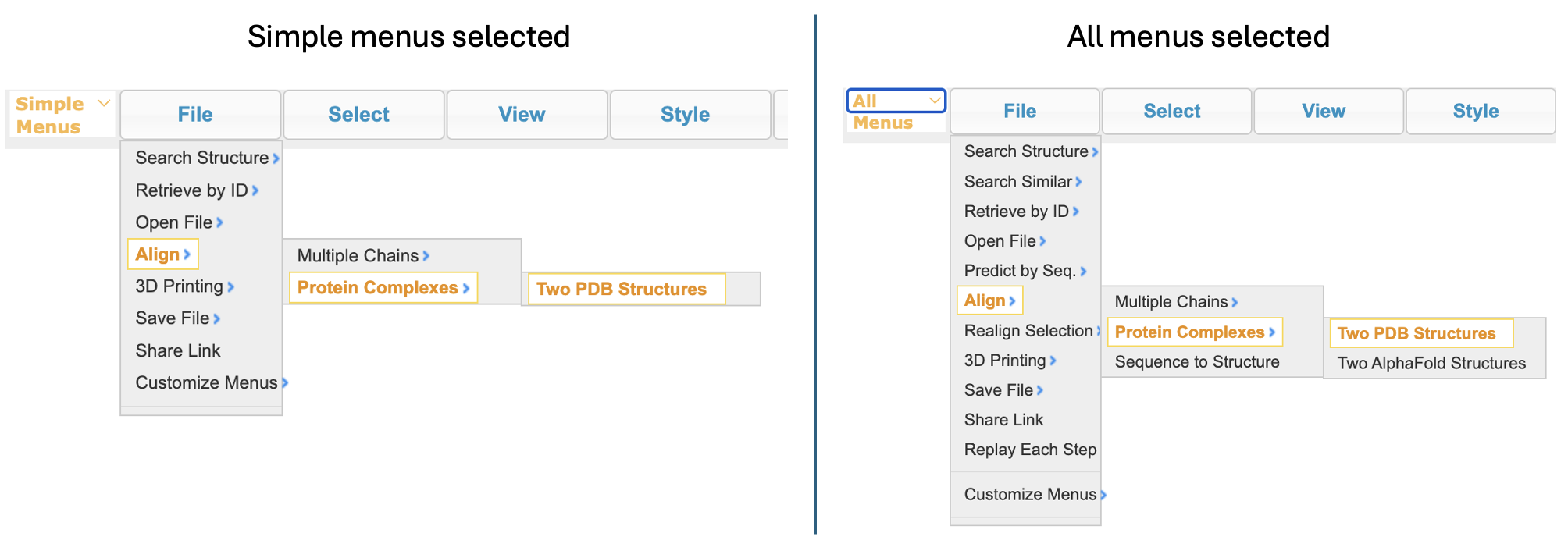
-
In the popup window:
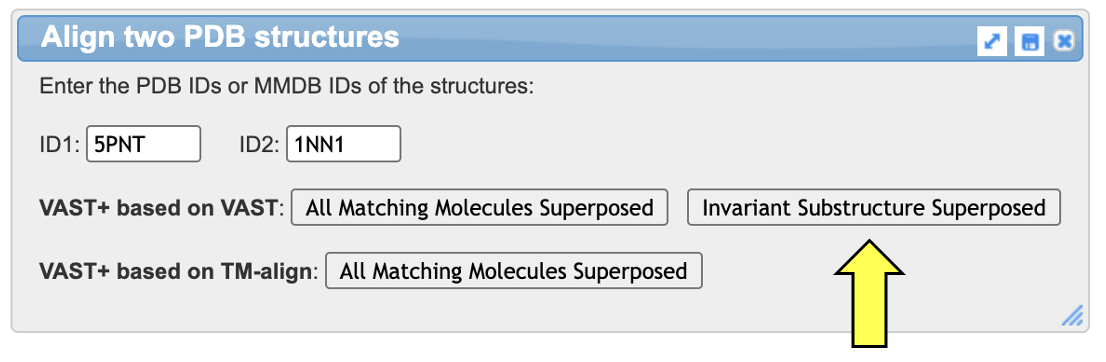
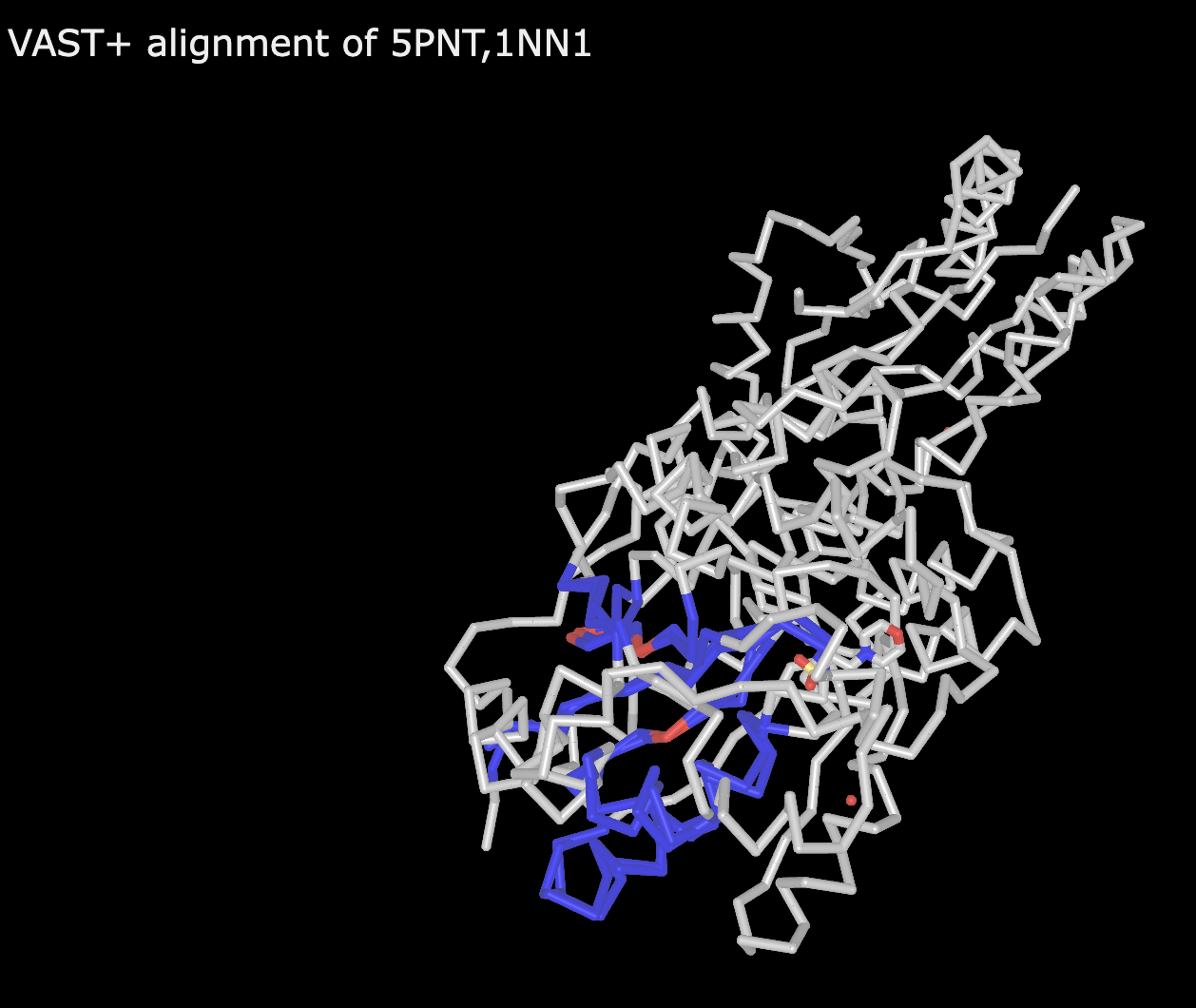
Note: Red regions indicate aligned residues with identical amino acids in the two structures, blue indicates aligned residues but non-identical amino acids, and gray indicates non-aligned residues. See example
- Repeat the above steps, this time using 5PNT and 6Y2V, both human phosphatases with 85% sequence ID
-
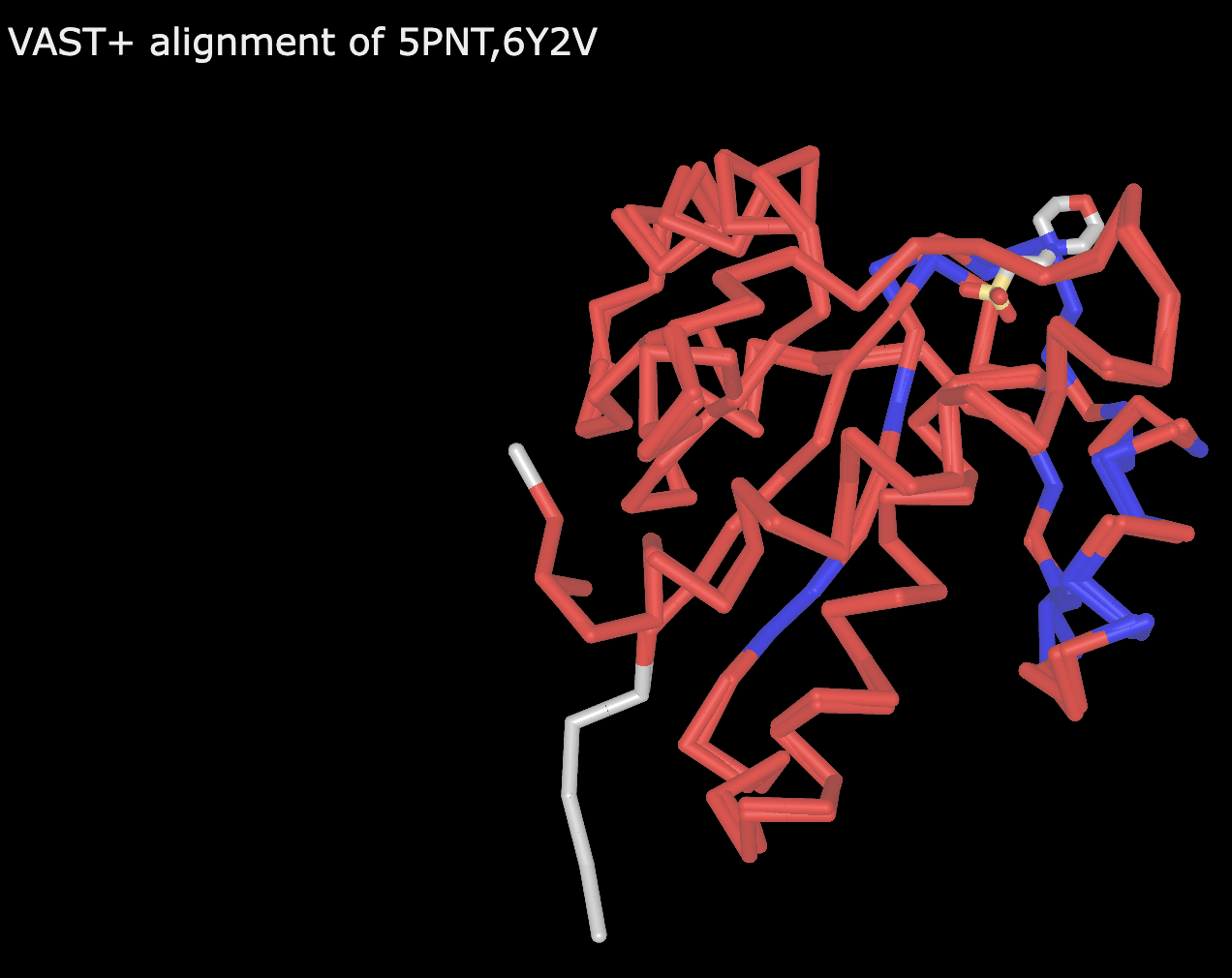
Figure 4: Alignment of 5PNT and 6Y2V With the trackpad or mouse, click anywhere within the structure viewer.
- Use the “a” key to toggle between the two aligned structures.
