XI.A. ChimeraX Superimposition
Walter Novak and Josh Beckham
Overview: This activity demonstrates how to align two objects in the modeling program.
Outcome: The user will be able to superimpose two similar macromolecules to examine commonalities and differences.
Time to complete: 5 minutes
Modeling Skills
- Superimposing objects
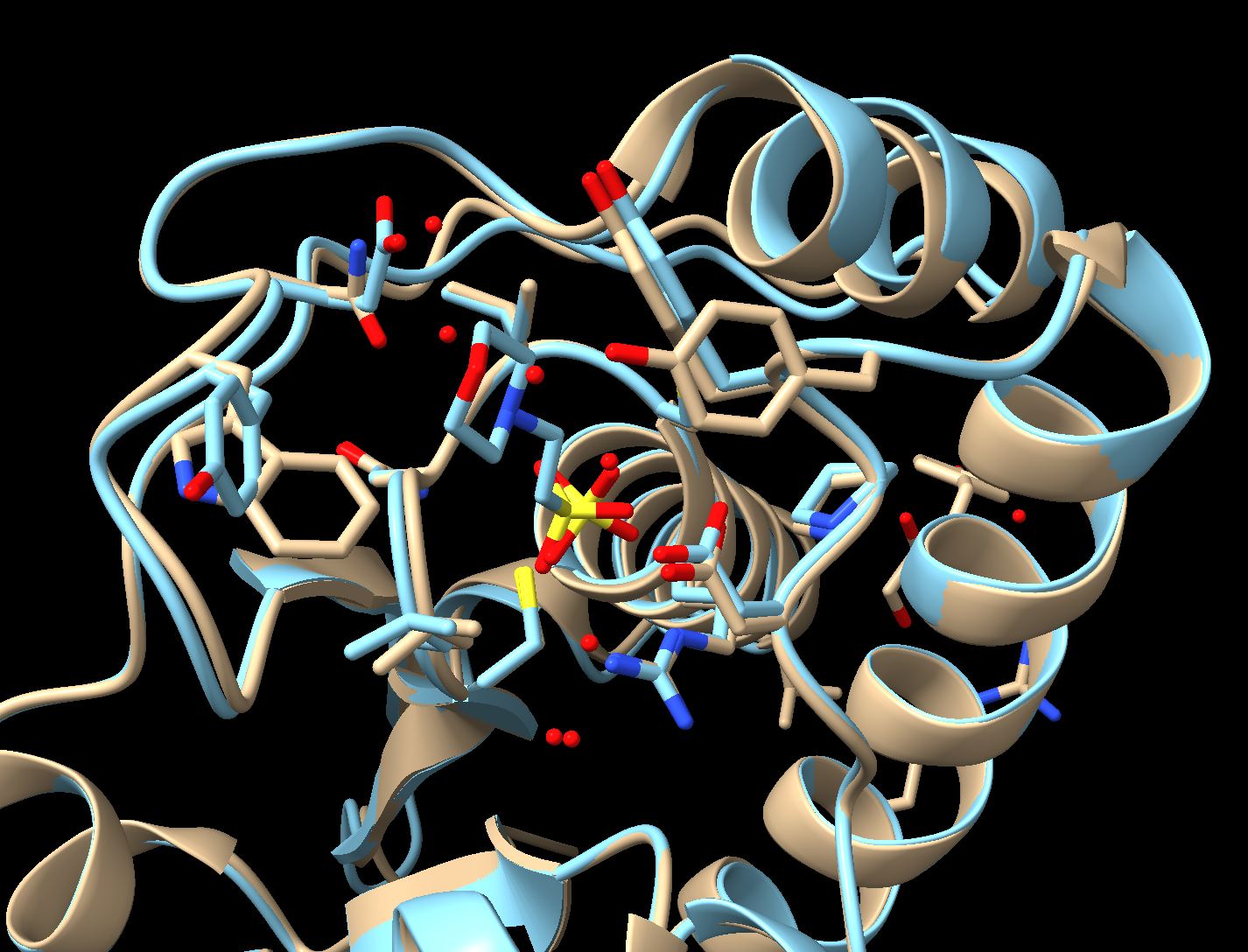
About the Models
PDB ID: 1xww & 5pnt (different isozymes)
Protein: Low molecular weight protein tyrosine phosphatase
Activity: hydrolyzes Tyr-OPO32- phosphoester bond
Description: single chain, bound SO42- competitive inhibitor, bound glycerol (nonspecific stabilizer)
Steps
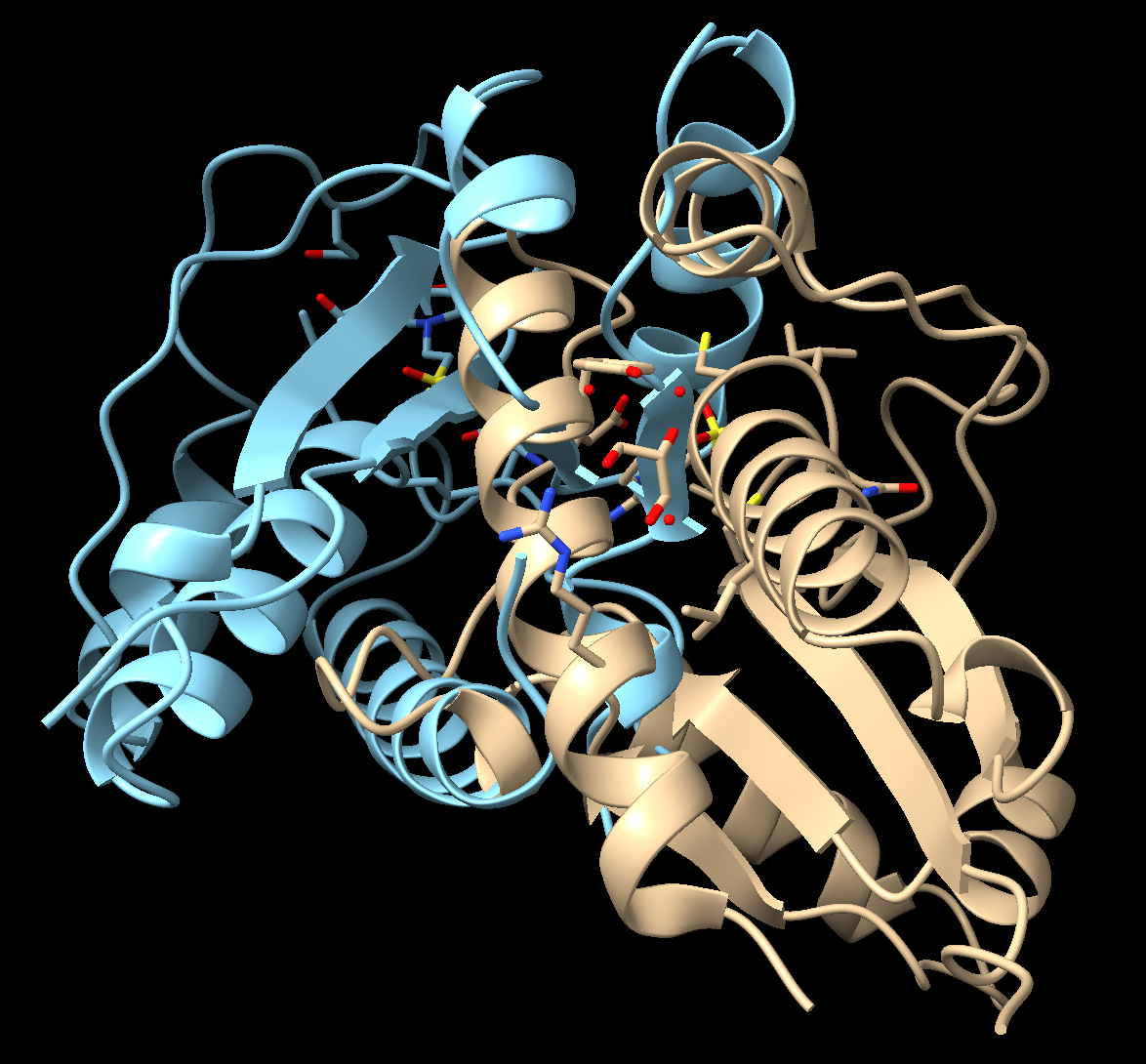
- Load two structures. In the command line, type: open 1xww 5pnt
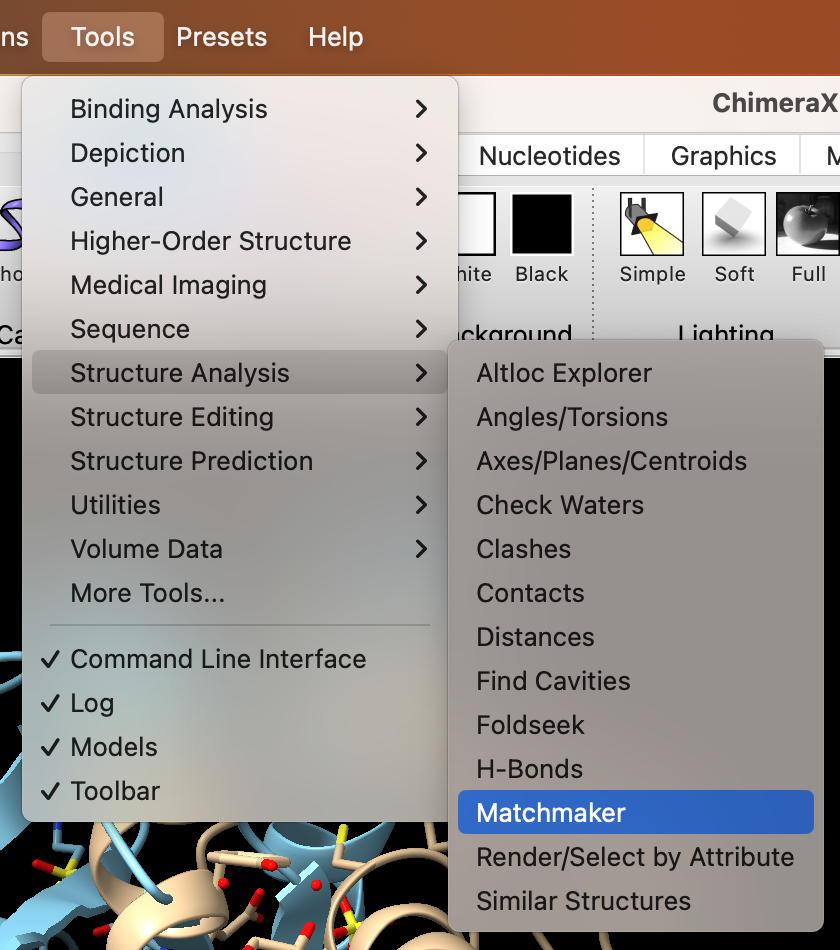
- Use the dropdown menu to superimpose the structures: Tools → Structure Analysis → Matchmaker (Figure 2).
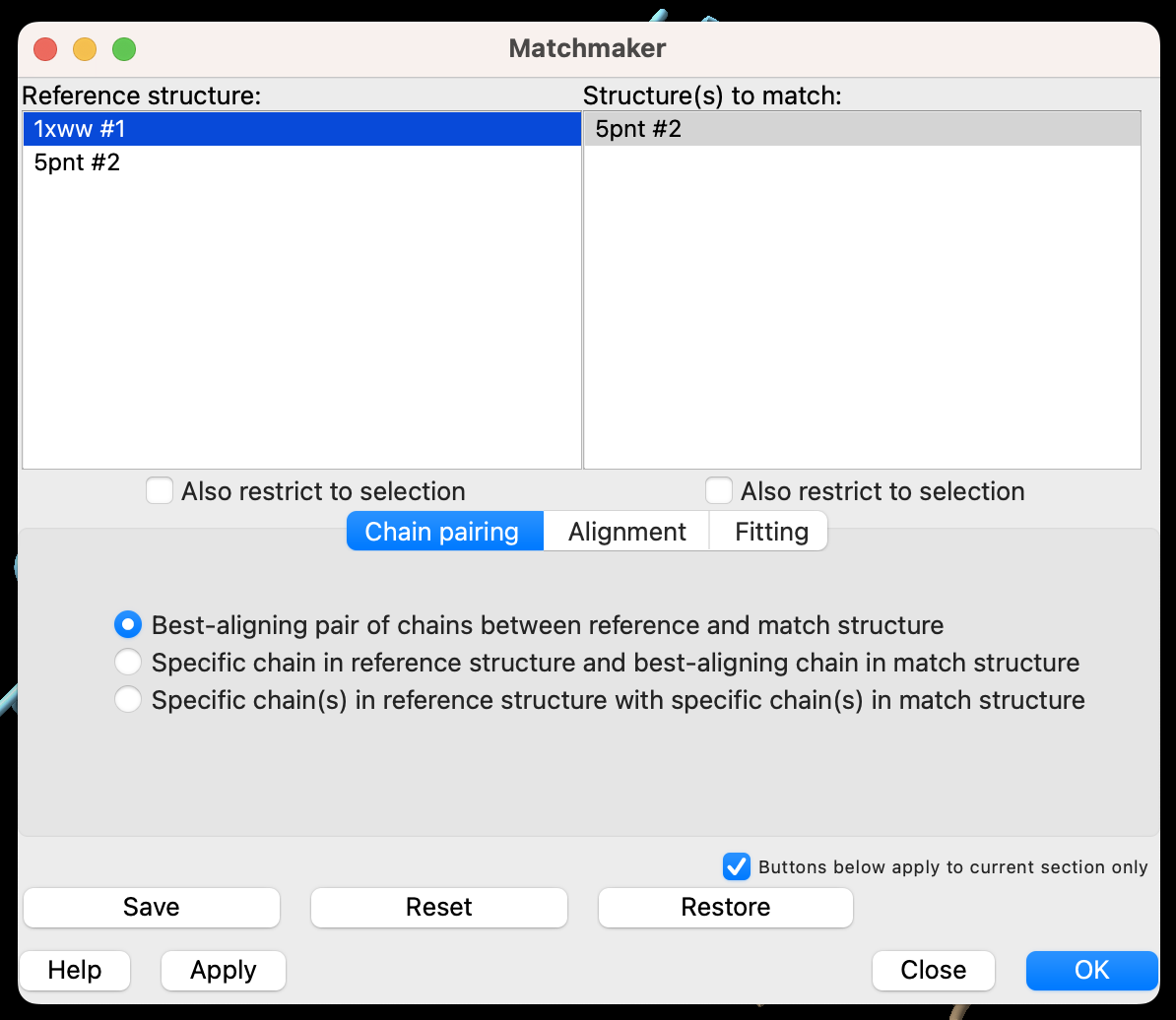
- You can control which structure is the reference and which will be moved to align in the popup window (Figure 3). The defaults are fine for this exercise. In the popup window, click “OK.”
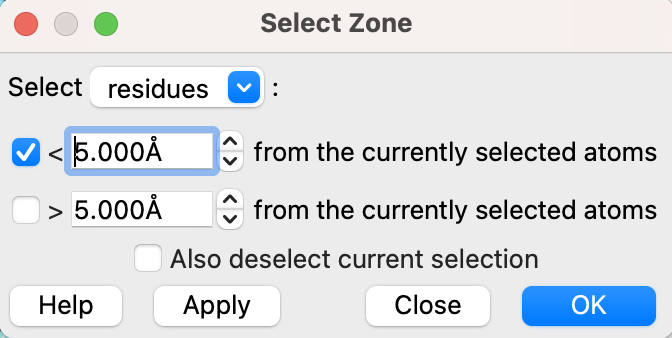
- Select a 5 Å zone around MES (2-(N-Morpholino)-ethanesulfonic acid) and display those residues (Figure 4).
a) Dropdown menu: Select → Residues → Mes
b) Dropdown menu: Select → Zone
c) In the popup window, set the selector to “residues” and “<5.000Å” from the currently selected atoms. Click “OK.”
d) Click “Show” in the “Atoms” section of the “Home” toolbar to show these residues.
- Reorient the proteins to identify differences between the active sites.
- (Optional) Save and close the session.
