IX.D. PyMOL Manual Measurement
Roderico Acevedo and Kristen Procko
Overview: This activity demonstrates how to manually measure distances.
Outcome: The user will be able to measure and label distances between two atoms (e.g., from an atom in a ligand to an atom in the enzyme active site).
Time to complete: 15 minutes
Modeling Skills
- Selecting objects using the command line
- Selecting groups within 5 Angstroms
- Manually measure distances between atoms (e.g., from a ligand to an active site amino acid residue)
About the Model
PDB ID: 1eve
Protein: Acetylcholinesterase
Activity: Hydrolyzes the neurotransmitter acetylcholine to form acetate and choline
Description: Single chain, bound to the drug Aricept (donepezil), covalently bound to five NAG glycan molecules
Steps
Use the Command Line to Show the Active Site Waters
- In the command line, type: fetch 1eve, type=pdb1
- Use the command line to select the active site:
| select aricept, resn E20 | %creates aricept as a selection |
| hide nonbonded | %hides waters |
| sele active, byres all within 5 of aricept | %finds residues within 5A of aricept |
| show sticks, active | %displays sidechains of selection as sticks |
| select active_water, ((aricept)around 3.3) and (resn HOH) | %creates active water selection and finds all waters within 3.3A of aricept |
| show spheres, active_water | %displays selected waters as non-bonded spheres |
| alter active_water, vdw=0.5 | %changes the VDW radius of selected waters to 0.5A |
| rebuild | %resets structural viewer displays with updated parameters |
| zoom active | %zooms display window to active selection |
- Color Aricept by element, with white carbons, so that it stands out. In the command line, type: util.cba(144,”aricept”,_self=cmd)
Note: If you receive the following error
SyntaxError: unterminated string literal (detected at line 1)
delete and re-type the quotation marks in the command util.cba(144,”aricept”,_self=cmd), as PyMOL will interpret both quotation marks as being the closing quotation marks. PyMOL requires both an open and close quotation mark.
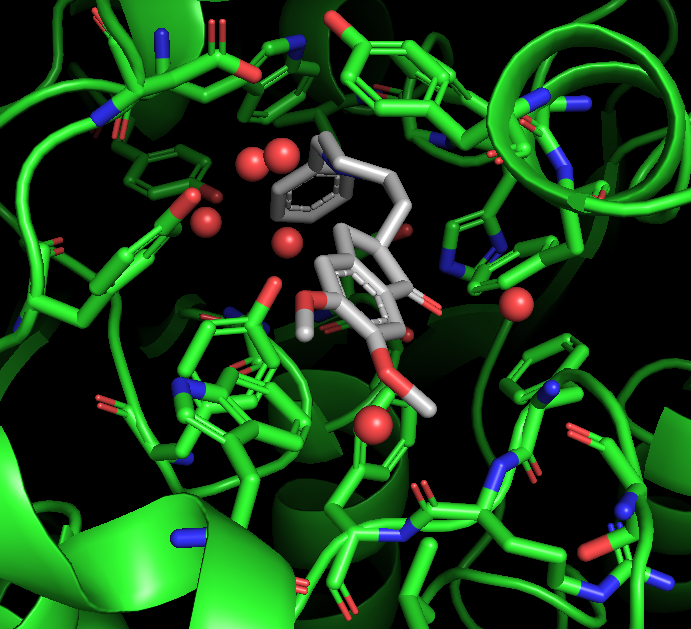
Find Polar Contacts
-
In the command line type: distance hbonds_active, aricept, active, 3.3, mode=2
Labeling
- Select into the background. This clears the selection.
-
To label the active site residues only (and not Aricept). In the names/object panel, click on “active”
- You should see both residues and Aricept highlighted (purple squares). Now, deselect Aricept by clicking on it. The purple squares should be removed from Aricept. Clear the selection by selecting into the background.
- Now label the active site residues by clicking: active → L → residues
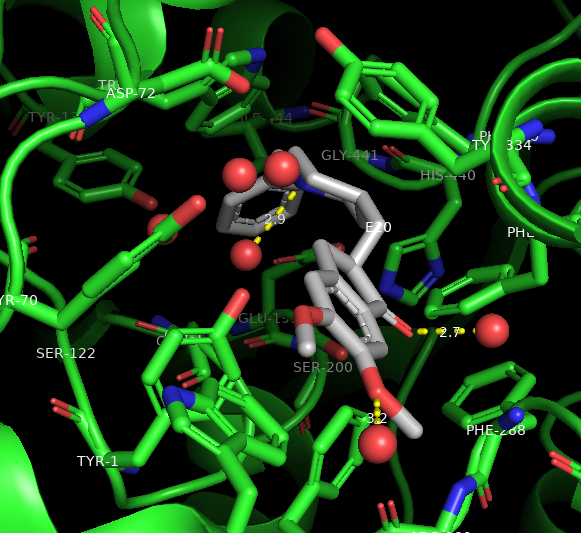
Figure 2: Results from finding polar contacts and labeling. Yellow dashed lines represent hydrogen bonds to active water molecules.
Note: If E20 appears multiple times, you did not deselect Aricept. You will need to hide the labels and retry deselecting Aricept from the active selection.
- Switch the mouse selection from residues to atoms (orange box)
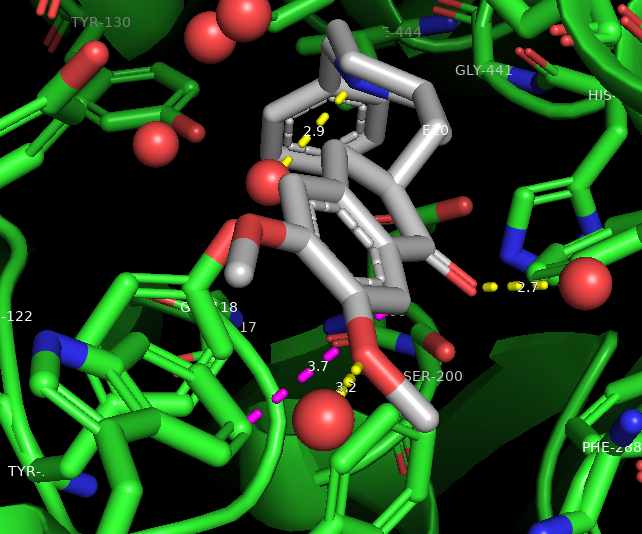
Figure 3: Using the measurement tool. The magenta dashed line represents a van der Waals interaction between Aricept (gray) and W-279 (green sticks). - Click on a carbon atom on Aricept. A new selection will appear in the names/object panel. Click sele → L → residue names
Use the Measurement Wizard to Find Interactions
- In the control panel, click on the wand icon, then select “measurement” from the dropdown menu
- Click on one of the aromatic rings of Aricept to see if you can find van der Waals interactions to any nearby aromatic residues (these measurements range from 3.8 – 4.2A)
- A distance is made by selecting any two atoms. When you’re finished measuring, click done.
- Unneeded measurements can be deleted, and useful measurements renamed, as appropriate
- You can also change the color of the dashed line by selecting the measurement → C and choosing a color (the image shows the dashed line as magenta).
Click here to go to Chapter X: Protein-Protein Interface
