XII.D. PyMOL Mutagenesis
Julie Himmelberger and Pamela Mertz
Overview: This activity demonstrates how to manually modify an amino acid residue to simulate protein mutagenesis.
Outcome: The user will be able to change the identity of an amino acid residue in order to visualize the impact of mutagenesis on the protein of interest.
Time to complete: 10 minutes
Modeling Skills
- Changing the identity of an amino acid residue using the mutagenesis tool
- Selecting the rotamer that best fits the structure
- Visualizing the impact of mutagenesis on the contour of a cavity in a protein
About the Models
PDB ID: 4LTV
Protein: Epi-isozizaene synthase (EIZS)
Activity: Cyclizes the isoprenoid precursor, farnesyl diphosphate (FPP) to form the tricyclic terpene epi-isozizaene. Epi-isozizaene is an intermediate in the biosynthesis of the natural product albaflavenoid.
Description: Single chain, no ligands bound in the active site, two sulfate ions
Note: We will be using the commands to change the identity of a specific amino acid residue in the enzyme. The amino acid residue, F96, that is mutated in this exercise is on the inner surface of the active site pocket. Mutagenesis experiments (Aaron et al, 2010, Li. et al., 2014) have shown that when this residue is mutated, the change in the contour and electrostatics of the active site pocket result in a dramatic change in the identity of isoprenoid products formed. The F96A mutation changes main product of the enzyme from epi-isozizaene to β–farnesene, and the F96W mutation changes the main product from epi-isozizaene to (+)-zizaene.
Steps
Substitution (mutagenesis) of an amino acid residue with rotamer selection
Note: This set of steps will produce a mutation at a single amino acid residue in the protein and allow the user to select a conformation of the new residue that has the fewest steric clashes with existing atoms in the structure.
- In the command line, type: fetch 4LTV, type=pdb1
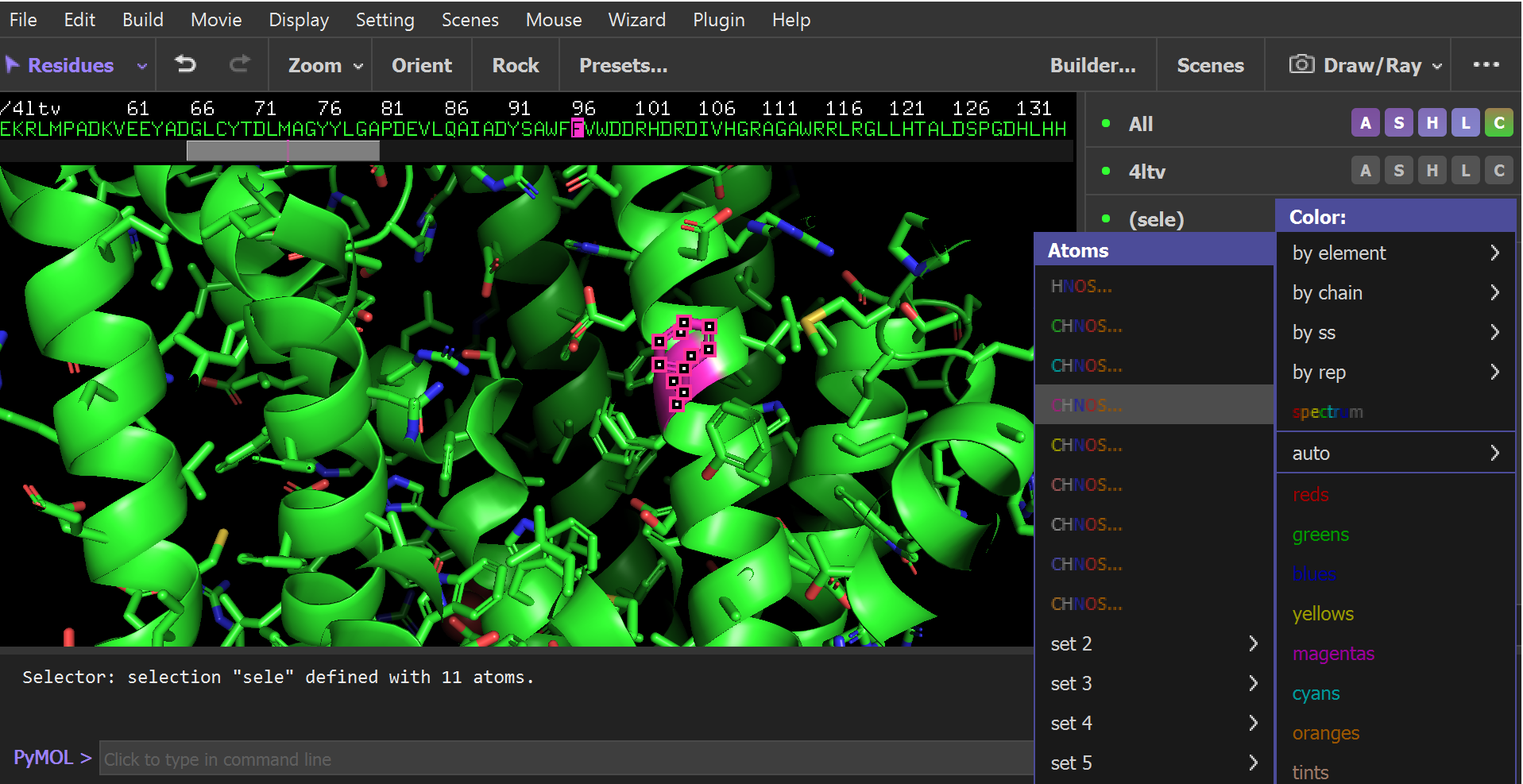
Figure 1: Changing the color of the amino acid residue that will be changed by mutagenesis. Residue F96 is colored magenta. - To hide the water molecules, type:
hide nonbonded - Next to 4LTV in the names/object panel, click: S → side chain → sticks.
- Display the amino acid sequence by clicking: Display → Sequence in the drop-down menu.
- In the sequence viewer at the top of the screen, click on residue F96. A new selection will appear in the names/object panel called “(sele)”.
- Next to “(sele)” click: C → by element → (option with magenta carbons). Next to “(sele)” click: A → center to center the amino acid in the screen. Zoom in so that you can easily see F96, then click in the open area of the structure viewer deselect everything.
- To mutate residue F96, in the drop-down menu click: Wizard → Mutagenesis → Protein.
- Click on F96 in the sequence viewer at the top of the screen. Then, in the mutagenesis menu that opens on the bottom right, click on: No mutation, then click on TRP.
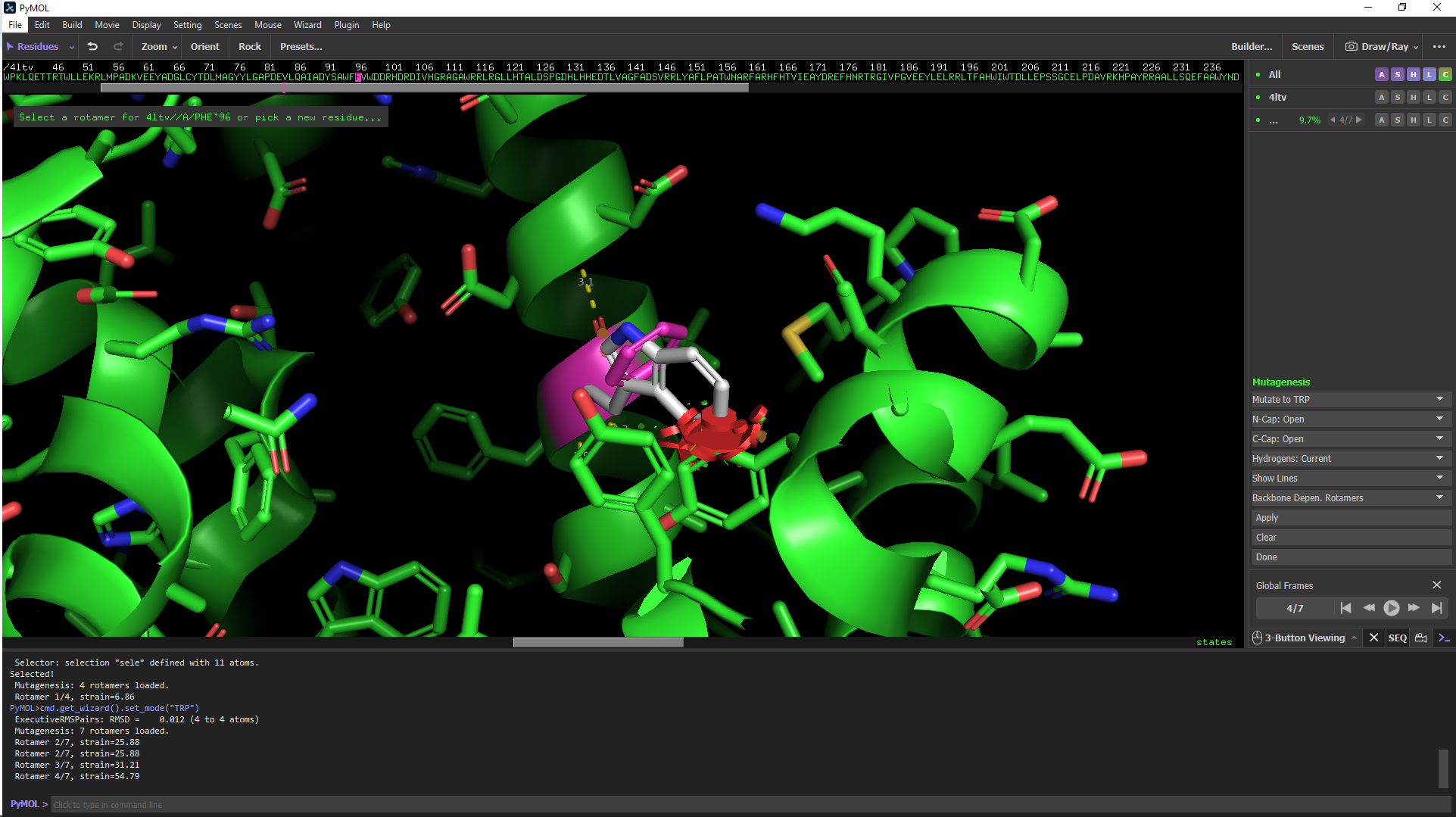
Figure 2. Selecting a Rotamer. There are 7 possible rotamers for the F96W mutation. Rotamer 4 results in significant steric clashes as indicated by the red surface.to open the dropdown menu to select the new amino acid, click on TRP. - For many residues, including TRP, there are several possible rotamers (common side chain conformations) that are possible. Toggle through the options by clicking on the forward and backward arrows under Global Frames (bottom right). For the F96W mutation, rotamer 2/7 has the least steric hindrance (indicated by red clashes), whereas rotamer 4/7 results in significant steric clashes with nearby side chains. See Figure 2.
- When you are viewing the preferred rotamer, click Apply, then Done.
- Save a session to keep the structure of the mutant.
Visualizing change in active site pocket due to mutagenesis
Note: This set of steps will produce a mutation at a single amino acid residue in the protein and allow the user to visualize the difference in volume of the active site pocket for the wild-type and mutant proteins.
-
In the command line, type: fetch 4LTV, type=pdb1
-
To hide the water molecules, type:
hide nonbonded -
Next to 4LTV in the names/object panel, click: S → side chain → sticks.
-
Display the amino acid sequence by clicking: Display → Sequence in the drop-down menu.
-
In the sequence viewer at the top of the screen, click on residue F96. A new selection will appear in the names/object panel called “(sele)”.
-
Next to “(sele)” click: C → by element → (option with magenta carbons). Next to “(sele)” click: A → center to center the amino acid in the screen. Zoom in so that you can easily see F96, then click in the open area of the structure viewer deselect everything.
- Hide side chains and show only the cartoon by clicking on (in the 4LTV row): H → side chain.
- Duplicate the 4LTV object by clicking on: A → copy to object → new.
- Redisplay F96 by clicking on that amino acid in the row for 4LTV in the sequence viewer then choose S → side chain → sticks. Click in the open area of the structure viewer to deselect everything.
- Rename the obj01 by clicking on: A → rename object and type the new name, F96A
- View only the new F96A object by clicking on the 4LTV object so that the name appears grey and it disappears.
- Mutate residue F96 to F96A by clicking: Wizard → Mutagenesis → Protein. Then click on the residue to be mutated (F96). In the mutagenesis menu that opens on the bottom right, click on: No mutation to open the dropdown menu and click on: Ala to select the new amino acid. Click on apply, then done. Alanine does not have any rotamers, so there is no rotamer selection.
- In the drop-down menu click on Setting → Surface → Wireframe.
- Click on Setting → Surface → Cavities & Pockets Only.
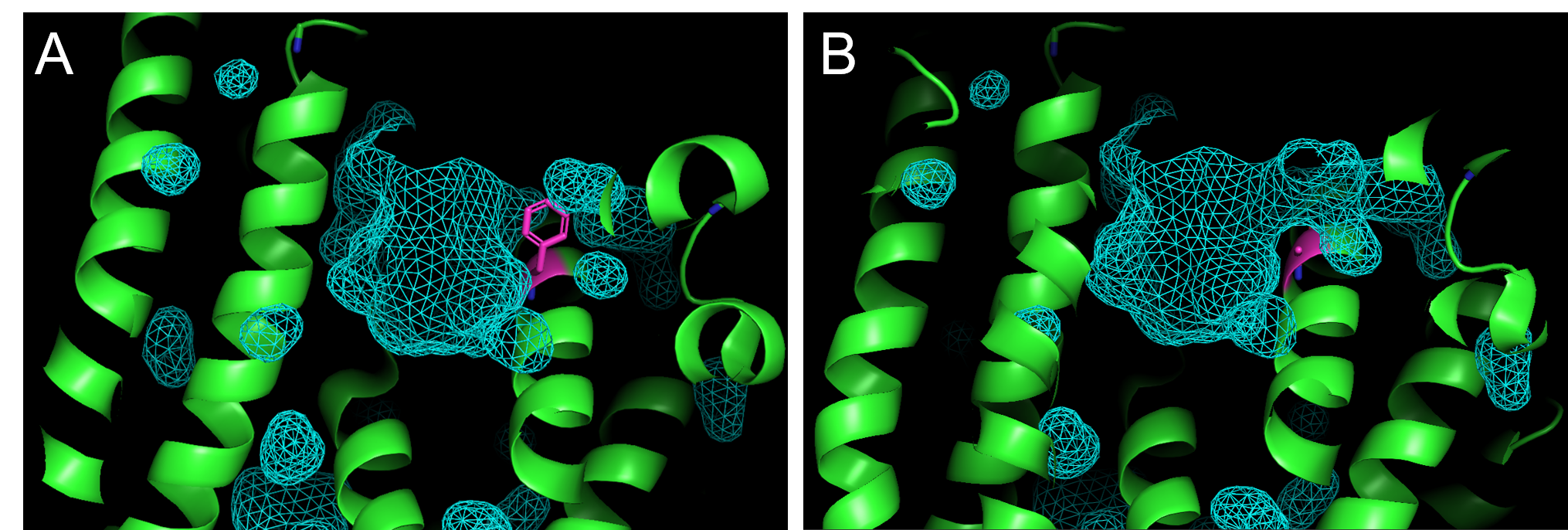
- Type in the command line: show surface and color the surface by typing in the command line:
set surface_color, cyan . A mesh outline of the active site should appear as shown in Figure 5. - You can toggle between the viewing the surface the active site cavity of the wild-type enzyme, 4ltv (Figure 4A) and the F96A mutant (Figure 4B) mutant by alternatively showing either 4LTV or F96A.