V.A. ChimeraX Mouse/Trackpad Selection
Walter Novak and Josh Beckham
Overview: This activity demonstrates how to select and color parts of a macromolecular structure.
Outcome: The user will be able to identify parts of a macromolecule of interest, select them with the mouse, and change the color and rendering of specific selections.
Time to complete: 15–20 minutes
Modeling Skills
- Selecting groups in the structure viewer window with the mouse
- Saving selections
- Rendering selections separately
About the Model
PDB ID: 6m0j
Protein: SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor
Activity: Angiotensin-converting enzyme 2 (Chain A); Spike protein S1 (Chain E); NAG – glycan; Zn2+; Cl–
Description: Receptor for the SARS-CoV-2 pandemic virus
Steps
Load the Structure
- In the command line: open 6m0j
Note: Notice in the log that chain A is ACE-2 and chain E is the Spike protein S1. The default render is ribbon (cartoon) with black background; Zn ion shown as sphere; glycans shown as sticks
- Color the two chains differently.
a) Use the dropdown menu: Select → Chains → Angiotensin-converting enzyme 2 (chain A) (Figure 1).
b) Give it a new color of your choice using the dropdown menu: Actions → Color → All options (ensure that ”Coloring applies to” all checkboxes) and click on a color. Alternatively, you can select directly from the dropdown menu.
c) Click “By Heteroatom” after you have colored the chain differently to apply CPK coloring.
d) Repeat steps a–c for the spike protein S1.
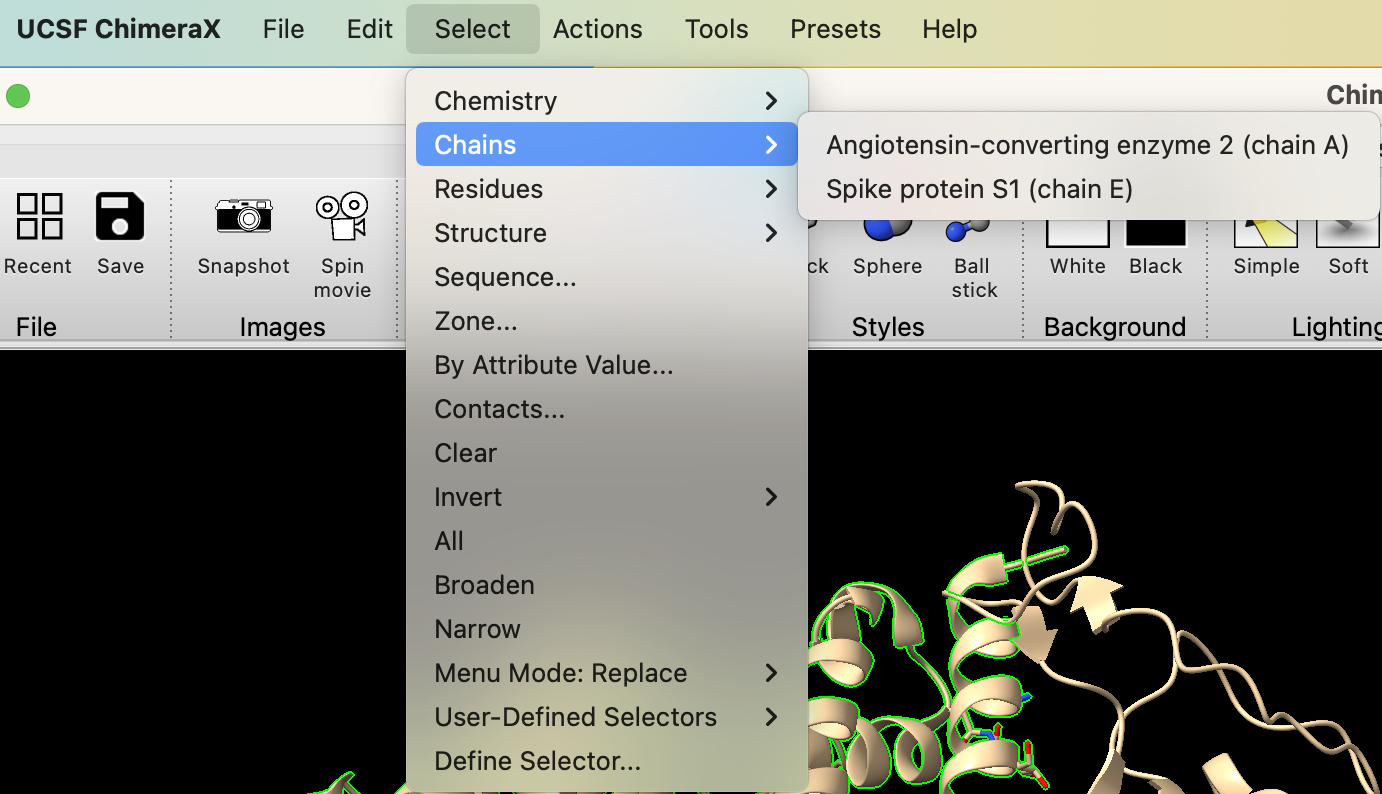
- Deselect the spike protein. In the dropdown menu: Select → Clear.
Select Glycans using the Mouse
- Notice the carbohydrates shown in stick rendering; these are covalently-bonded glycans. Using the mouse, orient the structure and use “Ctrl+Shift+Click” to select one bond or atom in each of the five glycans, which are distributed across the ACE-2 and Spike proteins.
NOTES
- It is useful to orient the structure so that the object you wish to select is not behind anything.
- If you mis-select, “Ctrl+Shift+Click” again on the selected part of the structure to deselect it.
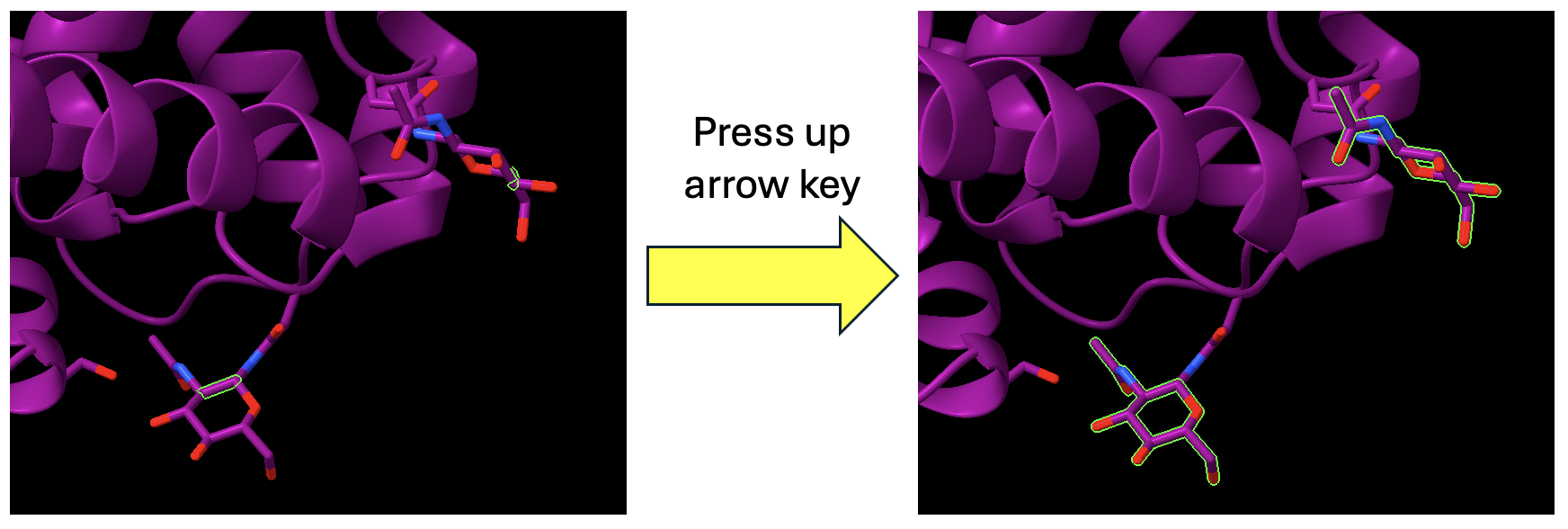
- To select all of the atoms and bonds in the glycans, broaden the selection by using the up arrow on your keyboard until all the atoms of the glycans are selected (Figure 2). If you hit the up arrow too many times, you will select the entire protein. No problem, simply click the down arrow until only the glycans are selected.
- Use the dropdown menu to define this selection.
a) Select → Define Selector.
b) Name the current selection “glycans” by typing in the textbox.
- “Ctrl+click” anywhere in the empty space of the structure viewer to clear the selection.
Rendering
- Color the Spike protein by secondary structure (colors both proteins at the start).
a) In the pulldown menu: Select → Structure → Secondary Structure → Helix.
b) Open the “Color Actions” panel: Actions → Color → All Options. Ensure only “Cartoons” is selected and click “Purple” to color the helices. Repeat for strands, but color them yellow.
- Color ACE-2 Plum with carbon atoms in Deep sky blue.
a) Use the pulldown menu to select ACE-2: Select → Chain → Angiotensin Converting Enzyme 2 (Chain A).
b) Use the “Color Actions” panel, Actions → Color → All Options. Click “Show all colors” to reveal more color options (Figure 3). Ensure only “Cartoons” is selected and click “Plum” to color the cartoon.
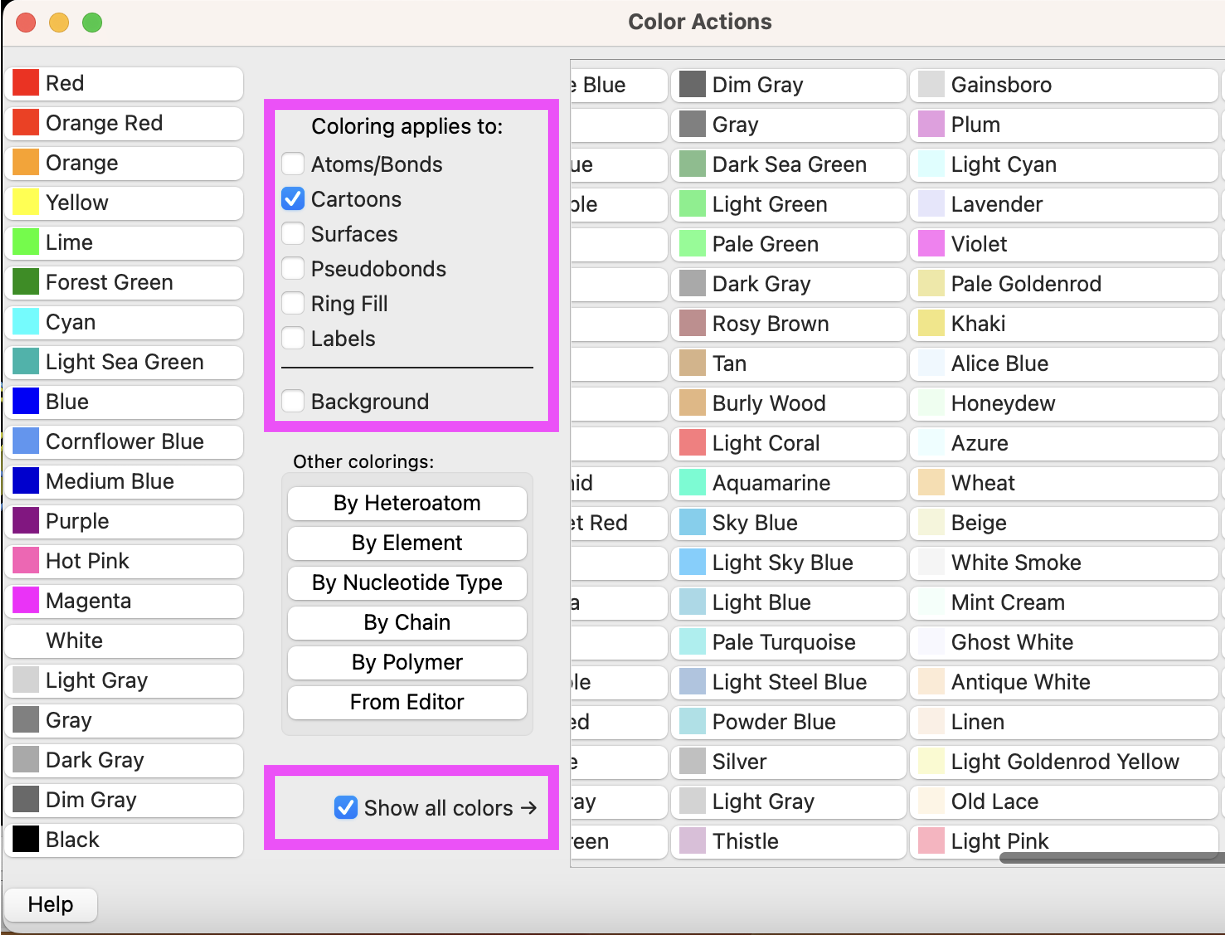
c) Then, change the “Coloring applies to:” selection to “Atoms/Bonds”, select “Deep Sky Blue”, then select “By Heteroatom.”
Note: There is no need to close out and reopen color window–you can make new selections without deselecting others (as they will automatically deselect.)
- Color the ions gray and the glycans as spheres with medium aquamarine carbons.
a) In the dropdown menu, “Select” → Structure → Ions.
b) Ensure that only Atoms/Bonds is checked in the “Coloring Applies to:” section of the “Color Actions” panel. Color them gray by clicking “Gray” in the panel.
c) Use the dropdown menu to select the glycans. Select → User-Defined Selectors → glycans.
d) In the “Color Actions” panel, Actions → Color → All Options. Ensure only Atoms/Bonds is selected and click “Medium Aquamarine” to color the atoms.
e) Click “By Heteroatom” in the “Color Actions” panel to color non-carbon atoms in CPK.
f) Close the “Color Actions” panel.
g) Use the ”Home” toolbar to click on “Sphere.”
Finishing Up
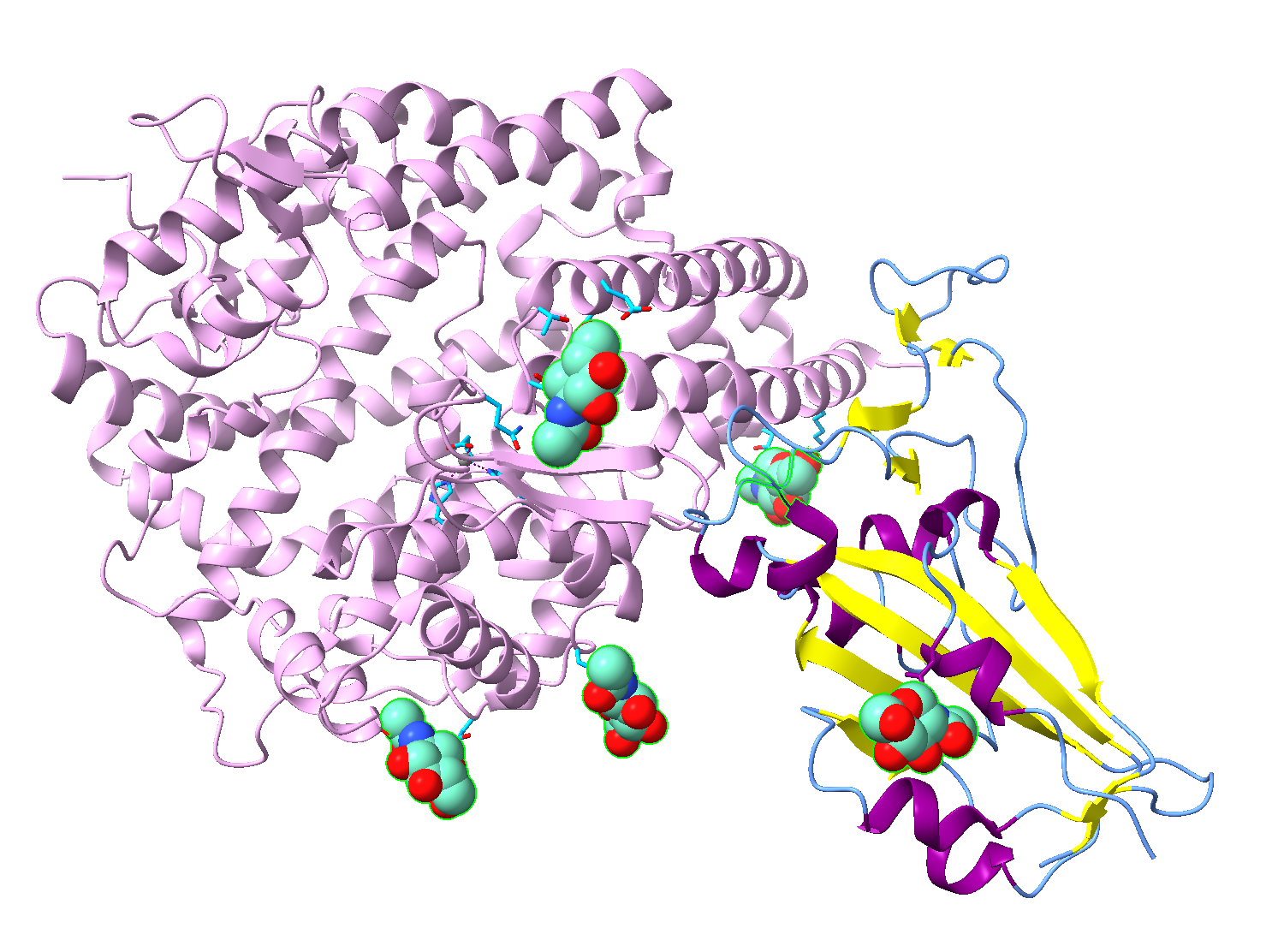
- Use the “Home” toolbar to change the background to white.
- (Optional) Save and close the session.
Jump to the next ChimeraX tutorial: VI.A. ChimeraX Selecting on the Sequence and Labeling.
