6. Advanced Search through the RCSB PDB
Walter Novak
Advanced searching is a powerful way to sift through the more than 200,000 structures available in the PDB. For example, you can quickly search for specific ligands or cofactors, enzyme classification (E.C.) numbers, experiment type, or even sequence and structural motifs. Further, individual advanced searches can be combined to yield highly focused results. The RCSB allows advanced searches using sequence information, structural information, chemical information or other characteristics called “attributes.”
To use the Advanced Search Query Builder (Figure 1), simply point your web browser to the RCSB PDB website and just under the search box click the “Advanced Search” link. It can be a little daunting at first, but our instructions below will have you searching like an expert in no time.
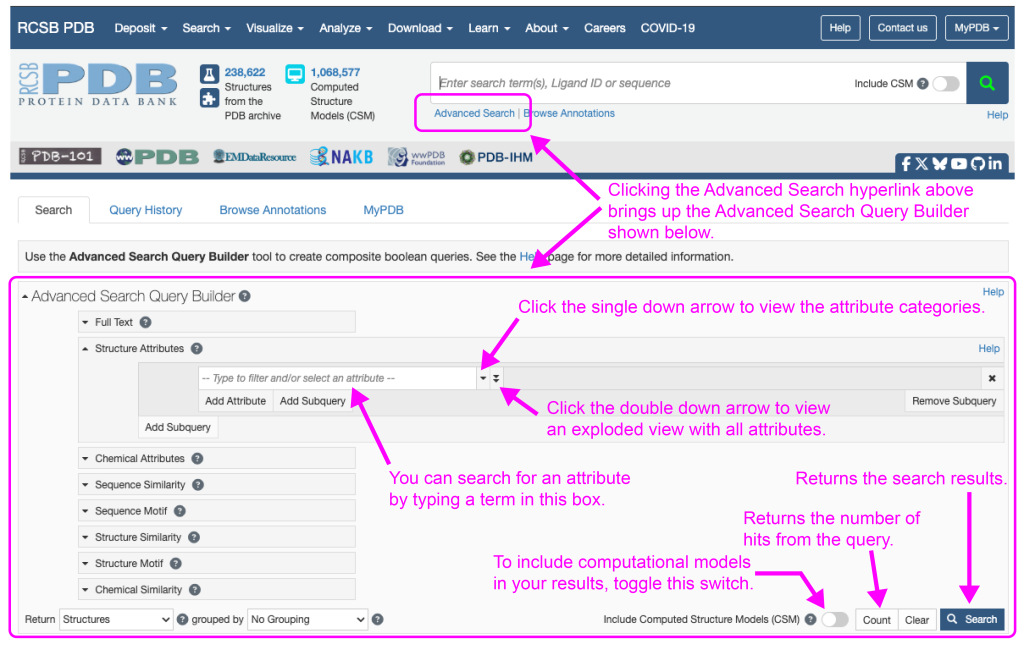
You can search for an attribute by typing in the “Type to filter and/or select an attribute” box. You can also identify categories of attributes or all attributes by clicking the single or double down arrows, respectively, at the right of the attribute box (Figure 1). In the bottom right hand corner of the Advanced Search Query Builder you can choose to include computationally derived structural models. You can also return the number of hits your query will generate with the “Count” button. Finally, clicking “Search” will return your search results.
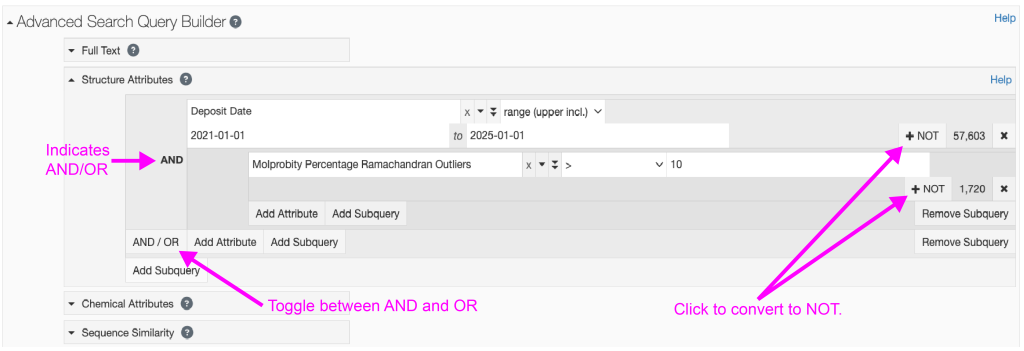
One final note is that you can use Boolean operators (AND, OR, or NOT) to yield results that include this AND that, this OR that, or this, but NOT that. Figure 2 below gives an example of how to use Boolean operators to identify structures with a deposit date from January 1, 2021 to January 1, 2025 that also have more than 10% of the protein residues identified as Ramachandran outliers by Molprobity.
We provide detailed instructions for several popular searches below.
Search Instructions available:
| Structure Similarity | PDB composition (Protein, NA, sugar) | |
| Sequence Similarity | PDB size | |
| Enzyme Classification Number |
Structure Similarity
There are two ways to identify structures that share a similar fold to a particular structure: 1) use an existing PDB ID and 2) upload your own PDB file. Here we will cover using an existing PDB ID.
Steps:
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
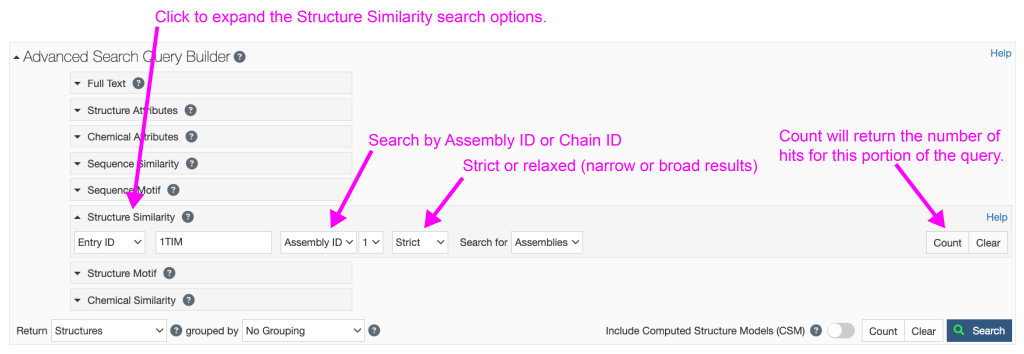
- Click on “Structure Similarity” under Advanced Search Query Builder to expand the menu (Figure 3).
- Type the PDB ID for your structure of interest (e.g. “1TIM” for triose phosphate isomerase) into the “Entry ID” search box and hit “Return.”
- You can search by “Assembly ID” or “Chain ID.”
- Assemblies may have multiple proteins or multiple copies of a single protein or nucleic acid. This is the default search. There may be multiple assemblies defined in your PDB, therefore you can select which assembly you want from the dropdown menu. If only one Assembly exists, then “1” will be your only choice.
-
- Note: To easily determine the biological assembly (assemblies) in your PDB, in another window navigate to the RCSB page for your structure of interest. On the left-hand side of the page you will see a cartoon of your structure. It will state the Biological Assembly at the top of this image. You can click through different assemblies (if they are present).
-
- Chain refers simply to a contiguous amino acid or nucleic acid sequence. If your protein has multiple chains, you can select which one to search with. This may be especially helpful if you are just interested in one particular protein chain among many.
- Assemblies may have multiple proteins or multiple copies of a single protein or nucleic acid. This is the default search. There may be multiple assemblies defined in your PDB, therefore you can select which assembly you want from the dropdown menu. If only one Assembly exists, then “1” will be your only choice.
- You can then select a “Strict” or “Relaxed” search. These choices make intuitive sense; strict will only find the most closely related structures, while relaxed will find all similar structures.
- Click “Count” on the far right to see how many hits meet your search criteria.
- To view the results, click the blue “Search” button on the far right.
Sequence Similarity
There are two ways to identify structures that share sequence similarity: 1) use an existing PDB ID and 2) paste in an amino acid sequence.
Steps using an existing PDB ID:
-
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
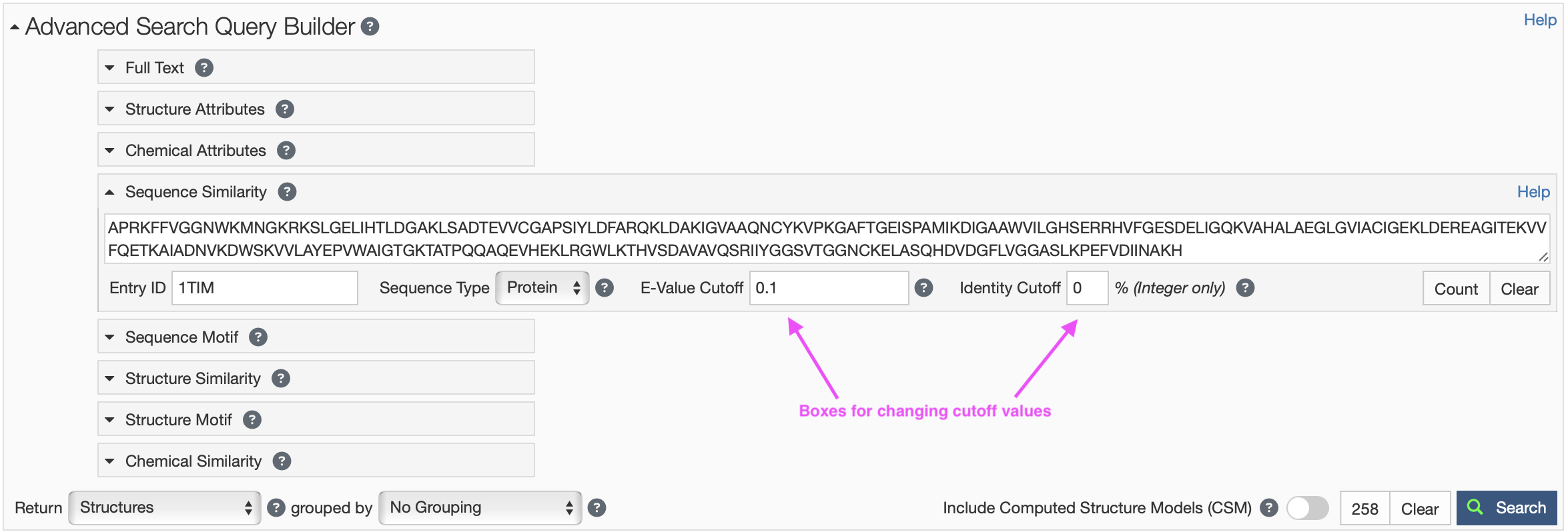
- Click on Sequence Similarity under the Advanced Search Query Builder to expand the menu to expand the menu (Figure 4).
- Type in the PDB ID you are interested in (e.g. “1TIM” for triose phosphate isomerase) in the “Entry ID” search box and hit “Return.” You should see the amino acid sequence appear in the sequence box.
- Click “Count” on the far right to see how many hits meet your search criteria.
- You can reduce the number of hits by either 1) reducing the E-value cutoff (e.g. “1E-100”) or increasing the Identity cutoff (e.g. 80).
- To view the results, click the blue “Search” button on the far right.
-
Steps using your own sequence:
-
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
- Click on Sequence Similarity under the Advanced Search Query Builder to expand the menu (Figure 4).
- Paste your amino acid sequence of interest in the sequence box.
- Click “Count” on the far right to see how many hits meet your search criteria.
- You can reduce the number of hits by either 1) reducing the E-value cutoff (e.g. “1E-100”) or increasing the Identity cutoff (e.g. 80).
- To view the results, click the blue “Search” button on the far right.
-
Enzyme Classification Number
To identify PDB entries for particular enzyme it can be helpful to search by the Enzyme Classification (EC) number since each enzyme has a unique EC number.
Steps:
-
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
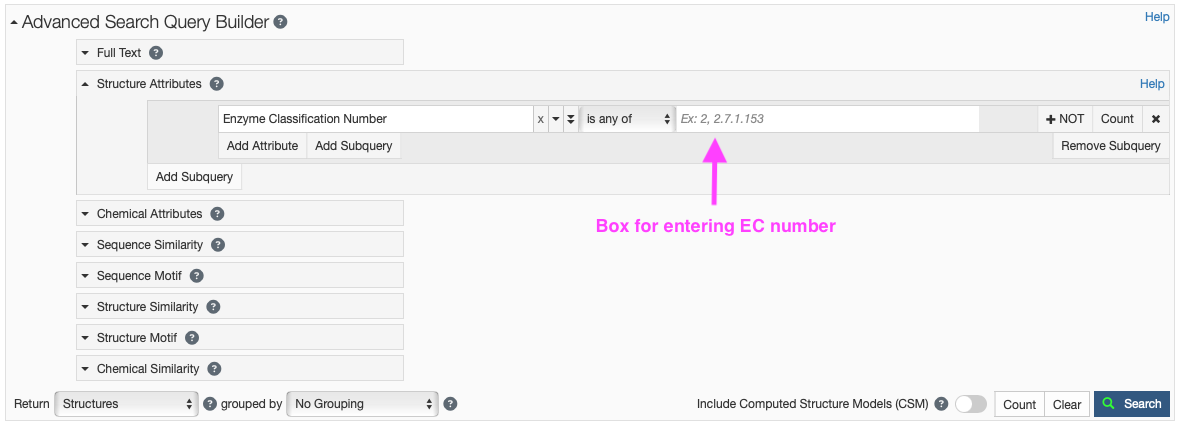
- Click on “Structure Attributes” under the Advanced Search Query Builder to expand the menu (Figure 5).
- Type “Enzyme” in the search box and you should see that under “Polymer Molecular Features,” two choices appear. Click “Enzyme Classification Number.”
- You can enter a whole or partial EC number (e.g. 2 or 2.1.3.2). You can also include several EC numbers in the search box, separated by commas.
- Your search will return hits containing any of the EC numbers in the search box.
- You may also choose to exclude certain EC numbers in your search by clicking the “+ NOT” button.
- Click “Count” on the far right to see how many hits meet your search criteria.
- To view the results, click the blue “Search” button on the far right.
-
PDB composition (Protein, NA, sugar)
PDB files have many other attributes that can be used to refine searches. Using the composition of the PDB file a search can be narrowed to a file that contains DNA, contains both protein and nucleic acid, or contains an oligosaccharide, for example.
Steps:
-
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
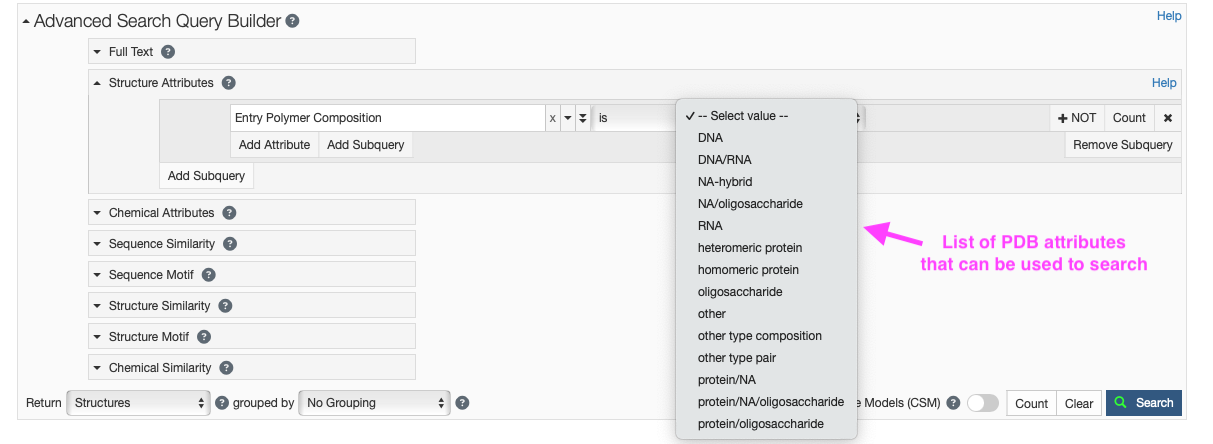
- Click on “Structure Attributes” under the Advanced Search Query Builder to expand the menu (Figure 6).
- Type “polymer” in the search box and you should click “Entry Polymer Composition” under “Entry Features.”
- Under the “– Select value –” pulldown you can select the composition of the PDB that you desire.
- You may also choose to exclude certain compositions in your search by clicking the “+ NOT” button.
- Click “Count” on the far right to see how many hits meet your search criteria.
- To view the results, click the blue “Search” button on the far right.
-
PDB size
PDB size can be searched two ways. The first method uses “polymer entity molecular weight” and will identify all PDB files that possess at least one chain with the given molecular weight. The second method uses “molecular weight per deposited model” and will only identify PDB files with that total molecular weight.
Steps using “Polymer Entity Molecular Weight”:
-
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
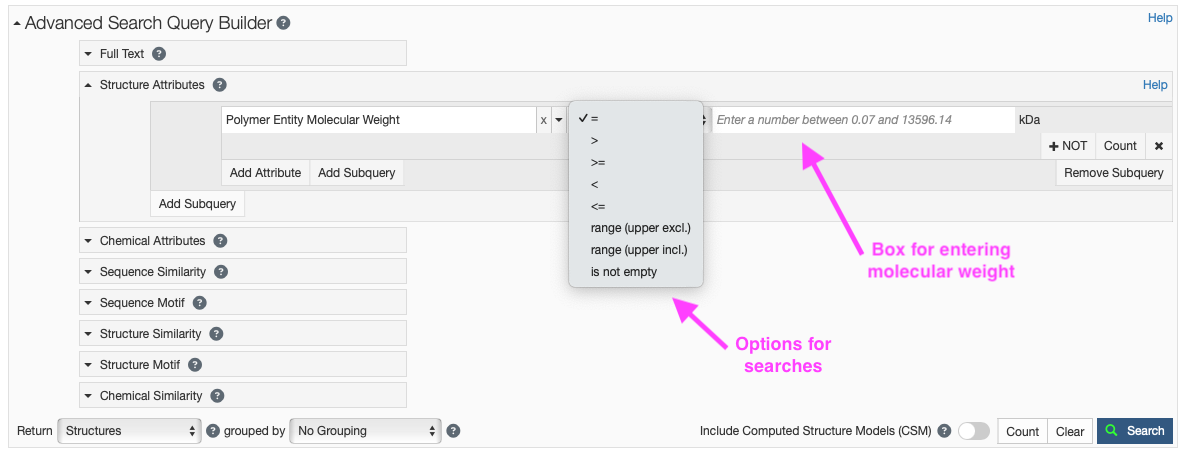
- Click on “Structure Attributes” under the Advanced Search Query Builder to expand the menu (Figure 7).
- Type “weight” in the search box and you should click “Polymer Entity Molecular Weight” under “Polymer Molecular Features.” The default is to search using the “=”; however, this dropdown menu can allow for a variety of searches (e.g. >, <, range).
- In the search box enter the kDa you would like to search on (e.g. 30).
- You may also choose to exclude certain compositions in your search by clicking the “+ NOT” button.
- Click “Count” on the far right to see how many hits meet your search criteria.
- To view the results, click the blue “Search” button on the far right.
-
Steps using “Molecular Weight per Deposited Model”:
-
-
- Under the search box at the RCSB, click on the “Advanced Search” link.
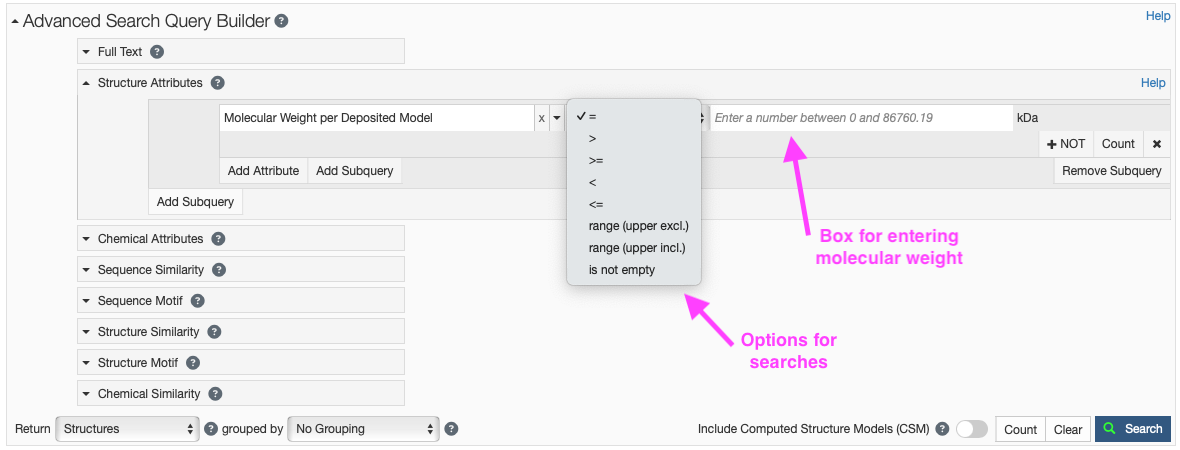
- Click on “Structure Attributes” under the Advanced Search Query Builder to expand the menu (Figure 8).
- Type “weight” in the search box and you should click “Molecular Weight per Deposited Model” under “Entry Features.”
- The default is to search using the “=”; however, this dropdown menu can allow for a variety of searches (e.g. >, <, range).
- In the search box enter the kDa you would like to search on (e.g. 30).
- You may also choose to exclude certain compositions in your search by clicking the “+ NOT” button.
- Click “Count” on the far right to see how many hits meet your search criteria.
- To view the results, click the blue “Search” button on the far right.
-
